Pericentromeric satellite lncRNAs are induced in cancer-associated fibroblasts and regulate their functions in lung tumorigenesis
- PMID: 36635266
- PMCID: PMC9837065
- DOI: 10.1038/s41419-023-05553-1
Pericentromeric satellite lncRNAs are induced in cancer-associated fibroblasts and regulate their functions in lung tumorigenesis
Abstract
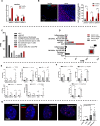
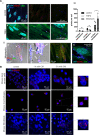
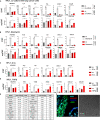
The abnormal tumor microenvironment (TME) often dictates the therapeutic response of cancer to chemo- and immuno-therapy. Aberrant expression of pericentromeric satellite repeats has been reported for epithelial cancers, including lung cancer. However, the transcription of tandemly repetitive elements in stromal cells of the TME has been unappreciated, limiting the optimal use of satellite transcripts as biomarkers or anti-cancer targets. We found that transcription of pericentromeric satellite DNA (satDNA) in mouse and human lung adenocarcinoma was observed in cancer-associated fibroblasts (CAFs). In vivo, lung fibroblasts expressed pericentromeric satellite repeats HS2/HS3 specifically in tumors. In vitro, transcription of satDNA was induced in lung fibroblasts in response to TGFβ, IL1α, matrix stiffness, direct contact with tumor cells and treatment with chemotherapeutic drugs. Single-cell transcriptome analysis of human lung adenocarcinoma confirmed that CAFs were the cell type with the highest number of satellite transcripts. Human HS2/HS3 pericentromeric transcripts were detected in the nucleus, cytoplasm, extracellularly and co-localized with extracellular vesicles in situ in human biopsies and activated fibroblasts in vitro. The transcripts were transmitted into recipient cells and entered their nuclei. Knock-down of satellite transcripts in human lung fibroblasts attenuated cellular senescence and blocked the formation of an inflammatory CAFs phenotype which resulted in the inhibition of their pro-tumorigenic functions. In sum, our data suggest that satellite long non-coding (lnc) RNAs are induced in CAFs, regulate expression of inflammatory genes and can be secreted from the cells, which potentially might present a new element of cell-cell communication in the TME.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

