Genomic atlas of the plasma metabolome prioritizes metabolites implicated in human diseases
- PMID: 36635386
- PMCID: PMC7614162
- DOI: 10.1038/s41588-022-01270-1
Genomic atlas of the plasma metabolome prioritizes metabolites implicated in human diseases
Abstract
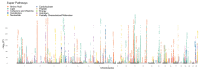
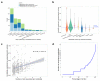
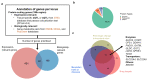
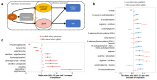
Metabolic processes can influence disease risk and provide therapeutic targets. By conducting genome-wide association studies of 1,091 blood metabolites and 309 metabolite ratios, we identified associations with 690 metabolites at 248 loci and associations with 143 metabolite ratios at 69 loci. Integrating metabolite-gene and gene expression information identified 94 effector genes for 109 metabolites and 48 metabolite ratios. Using Mendelian randomization (MR), we identified 22 metabolites and 20 metabolite ratios having estimated causal effect on 12 traits and diseases, including orotate for estimated bone mineral density, α-hydroxyisovalerate for body mass index and ergothioneine for inflammatory bowel disease and asthma. We further measured the orotate level in a separate cohort and demonstrated that, consistent with MR, orotate levels were positively associated with incident hip fractures. This study provides a valuable resource describing the genetic architecture of metabolites and delivers insights into their roles in common diseases, thereby offering opportunities for therapeutic targets.
© 2023. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Figures


References
-
- Bar N, et al. A reference map of potential determinants for the human serum metabolome. Nature. 2020;588 - PubMed
-
- Lee W-J, Hase K. Gut microbiota–generated metabolites in animal health and disease. Nat Chem Biol. 2014;10:416–424. - PubMed
-
- Wishart DS. Emerging applications of metabolomics in drug discovery and precision medicine. Nat Rev Drug Discov. 2016;15 - PubMed
-
- Long T, et al. Whole-genome sequencing identifies common-to-rare variants associated with human blood metabolites. Nat Genet. 2017;49 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

