A single-cell massively parallel reporter assay detects cell-type-specific gene regulation
- PMID: 36635387
- PMCID: PMC9931678
- DOI: 10.1038/s41588-022-01278-7
A single-cell massively parallel reporter assay detects cell-type-specific gene regulation
Abstract
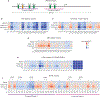
Massively parallel reporter gene assays are key tools in regulatory genomics but cannot be used to identify cell-type-specific regulatory elements without performing assays serially across different cell types. To address this problem, we developed a single-cell massively parallel reporter assay (scMPRA) to measure the activity of libraries of cis-regulatory sequences (CRSs) across multiple cell types simultaneously. We assayed a library of core promoters in a mixture of HEK293 and K562 cells and showed that scMPRA is a reproducible, highly parallel, single-cell reporter gene assay that detects cell-type-specific cis-regulatory activity. We then measured a library of promoter variants across multiple cell types in live mouse retinas and showed that subtle genetic variants can produce cell-type-specific effects on cis-regulatory activity. We anticipate that scMPRA will be widely applicable for studying the role of CRSs across diverse cell types.
© 2023. The Author(s), under exclusive licence to Springer Nature America, Inc.
Figures







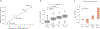
Comment in
-
A scalable technology for measuring cell-type-specific activity of cis-regulatory sequences.Nat Genet. 2023 Feb;55(2):172-173. doi: 10.1038/s41588-022-01277-8. Nat Genet. 2023. PMID: 36639574 No abstract available.
-
Consequences of cis-regulatory sequence variation.Nat Biotechnol. 2023 Feb;41(2):194. doi: 10.1038/s41587-023-01692-0. Nat Biotechnol. 2023. PMID: 36792710 No abstract available.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

