PhenoApp: A mobile tool for plant phenotyping to record field and greenhouse observations
- PMID: 36636476
- PMCID: PMC9813448
- DOI: 10.12688/f1000research.74239.2
PhenoApp: A mobile tool for plant phenotyping to record field and greenhouse observations
Abstract
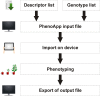
With the ongoing cost decrease of genotyping and sequencing technologies, accurate and fast phenotyping remains the bottleneck in the utilizing of plant genetic resources for breeding and breeding research. Although cost-efficient high-throughput phenotyping platforms are emerging for specific traits and/or species, manual phenotyping is still widely used and is a time- and money-consuming step. Approaches that improve data recording, processing or handling are pivotal steps towards the efficient use of genetic resources and are demanded by the research community. Therefore, we developed PhenoApp, an open-source Android app for tablets and smartphones to facilitate the digital recording of phenotypical data in the field and in greenhouses. It is a versatile tool that offers the possibility to fully customize the descriptors/scales for any possible scenario, also in accordance with international information standards such as MIAPPE (Minimum Information About a Plant Phenotyping Experiment) and FAIR (Findable, Accessible, Interoperable, and Reusable) data principles. Furthermore, PhenoApp enables the use of pre-integrated ready-to-use BBCH (Biologische Bundesanstalt für Land- und Forstwirtschaft, Bundessortenamt und CHemische Industrie) scales for apple, cereals, grapevine, maize, potato, rapeseed and rice. Additional BBCH scales can easily be added. The simple and adaptable structure of input and output files enables an easy data handling by either spreadsheet software or even the integration in the workflow of laboratory information management systems (LIMS). PhenoApp is therefore a decisive contribution to increase efficiency of digital data acquisition in genebank management but also contributes to breeding and breeding research by accelerating the labour intensive and time-consuming acquisition of phenotyping data.
Keywords: Android app; BBCH; FAIR principles; LIMS; digital data acquisition; plant phenotyping.
Copyright: © 2022 Röckel F et al.
Conflict of interest statement
No competing interests were disclosed.
Figures


References
-
- Norton SL, Khoury CK, Sosa CC, et al. : Priorities for enhancing the ex situ conservation and use of Australian crop wild relatives. Aust. J. Bot. 2017;65(8):638–645. 10.1071/BT16236 - DOI
-
- Pilling D, Bélanger J, Diulgheroff S, et al. : Global status of genetic resources for food and agriculture: challenges and research needs. Genet. Res. 2020;1(1):4–16.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

