The oral cancer microbiome contains tumor space-specific and clinicopathology-specific bacteria
- PMID: 36636719
- PMCID: PMC9831678
- DOI: 10.3389/fcimb.2022.942328
The oral cancer microbiome contains tumor space-specific and clinicopathology-specific bacteria
Abstract
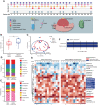
The crosstalk between the oral microbiome and oral cancer has yet to be characterized. This study recruited 218 patients for clinicopathological data analysis. Multiple types of specimens were collected from 27 patients for 16S rRNA gene sequencing, including 26 saliva, 16 swabs from the surface of tumor tissues, 16 adjacent normal tissues, 22 tumor outer tissue, 22 tumor inner tissues, and 10 lymph nodes. Clinicopathological data showed that the pathogenic bacteria could be frequently detected in the oral cavity of oral cancer patients, which was positively related to diabetes, later T stage of the tumor, and the presence of cervical lymphatic metastasis. Sequencing data revealed that compared with adjacent normal tissues, the microbiome of outer tumor tissues had a greater alpha diversity, with a larger proportion of Fusobacterium, Prevotella, and Porphyromonas, while a smaller proportion of Streptococcus. The space-specific microbiome, comparing outer tumor tissues with inner tumor tissues, suggested minor differences in diversity. However, Fusobacterium, Neisseria, Porphyromonas, and Alloprevotella were more abundant in outer tumor tissues, while Prevotella, Selenomonas, and Parvimonas were enriched in inner tumor tissues. Clinicopathology-specific microbiome analysis found that the diversity was markedly different between negative and positive extranodal extensions, whereas the diversity between different T-stages and N-stages was slightly different. Gemella and Bacillales were enriched in T1/T2-stage patients and the non-lymphatic metastasis group, while Spirochaetae and Flavobacteriia were enriched in the extranodal extension negative group. Taken together, high-throughput DNA sequencing in combination with clinicopathological features facilitated us to characterize special patterns of oral tumor microbiome in different disease developmental stages.
Keywords: 16S rDNA sequencing; clinicopathology; intratumoral bacteria; oral cancer (OC); oral microbiome.
Copyright © 2022 Zeng, Tan, Guo, Li, Yang, Lao, Wang, Ma, Zhang, Liao and Liang.
Conflict of interest statement
The authors declare that the research was conducted without any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

