Upregulation of PTPRC and Interferon Response Pathways in HIV-1 Seroconverters Prior to Infection
- PMID: 36637125
- PMCID: PMC9978315
- DOI: 10.1093/infdis/jiac498
Upregulation of PTPRC and Interferon Response Pathways in HIV-1 Seroconverters Prior to Infection
Abstract
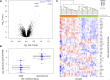
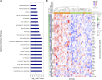
Human immunodeficiency virus 1 (HIV-1) exposed seronegative (HESN) individuals may have unique characteristics that alter susceptibility to HIV-1 infection. However, identifying truly exposed HESN is challenging. We utilized stored data and biospecimens from HIV-1 serodifferent couple cohorts, in which couples' HIV-1 exposures were quantified based on unprotected sex frequency and viral load of the partner with HIV-1. We compared peripheral blood gene expression between 15 HESN and 18 seroconverters prior to infection. We found PTPRC (encoding CD45 antigen) and interferon-response pathways had significantly higher expression among individuals who went on to become seropositive and thus may be a signature for increased acquisition risk.
Keywords: HIV-1; acquisition; gene expression; host resistance; microarray.
© The Author(s) 2023. Published by Oxford University Press on behalf of Infectious Diseases Society of America.
Conflict of interest statement
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

