Dealing with dimensionality: the application of machine learning to multi-omics data
- PMID: 36637211
- PMCID: PMC9907220
- DOI: 10.1093/bioinformatics/btad021
Dealing with dimensionality: the application of machine learning to multi-omics data
Abstract
Motivation: Machine learning (ML) methods are motivated by the need to automate information extraction from large datasets in order to support human users in data-driven tasks. This is an attractive approach for integrative joint analysis of vast amounts of omics data produced in next generation sequencing and other -omics assays. A systematic assessment of the current literature can help to identify key trends and potential gaps in methodology and applications. We surveyed the literature on ML multi-omic data integration and quantitatively explored the goals, techniques and data involved in this field. We were particularly interested in examining how researchers use ML to deal with the volume and complexity of these datasets.
Results: Our main finding is that the methods used are those that address the challenges of datasets with few samples and many features. Dimensionality reduction methods are used to reduce the feature count alongside models that can also appropriately handle relatively few samples. Popular techniques include autoencoders, random forests and support vector machines. We also found that the field is heavily influenced by the use of The Cancer Genome Atlas dataset, which is accessible and contains many diverse experiments.
Availability and implementation: All data and processing scripts are available at this GitLab repository: https://gitlab.com/polavieja_lab/ml_multi-omics_review/ or in Zenodo: https://doi.org/10.5281/zenodo.7361807.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2023. Published by Oxford University Press.
Figures
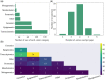

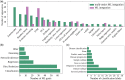
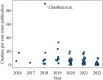

References
-
- Bahdanau D. et al. (2014) Neural machine translation by jointly learning to align and translate. arXiv, arXiv:1409.0473, preprint: not peer reviewed.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

