Concentration-Dependent Domain Evolution in Reaction-Diffusion Systems
- PMID: 36637542
- PMCID: PMC9839823
- DOI: 10.1007/s11538-022-01115-2
Concentration-Dependent Domain Evolution in Reaction-Diffusion Systems
Abstract
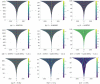
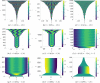
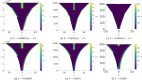
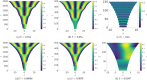
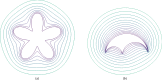
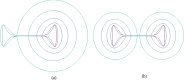
Pattern formation has been extensively studied in the context of evolving (time-dependent) domains in recent years, with domain growth implicated in ameliorating problems of pattern robustness and selection, in addition to more realistic modelling in developmental biology. Most work to date has considered prescribed domains evolving as given functions of time, but not the scenario of concentration-dependent dynamics, which is also highly relevant in a developmental setting. Here, we study such concentration-dependent domain evolution for reaction-diffusion systems to elucidate fundamental aspects of these more complex models. We pose a general form of one-dimensional domain evolution and extend this to N-dimensional manifolds under mild constitutive assumptions in lieu of developing a full tissue-mechanical model. In the 1D case, we are able to extend linear stability analysis around homogeneous equilibria, though this is of limited utility in understanding complex pattern dynamics in fast growth regimes. We numerically demonstrate a variety of dynamical behaviours in 1D and 2D planar geometries, giving rise to several new phenomena, especially near regimes of critical bifurcation boundaries such as peak-splitting instabilities. For sufficiently fast growth and contraction, concentration-dependence can have an enormous impact on the nonlinear dynamics of the system both qualitatively and quantitatively. We highlight crucial differences between 1D evolution and higher-dimensional models, explaining obstructions for linear analysis and underscoring the importance of careful constitutive choices in defining domain evolution in higher dimensions. We raise important questions in the modelling and analysis of biological systems, in addition to numerous mathematical questions that appear tractable in the one-dimensional setting, but are vastly more difficult for higher-dimensional models.
Keywords: Evolving domains; Linear instability analysis; Pattern formation.
© 2023. The Author(s).
Figures






References
-
- Baker RE, Maini PK. A mechanism for morphogen-controlled domain growth. J Math Biol. 2007;54(5):597–622. - PubMed
-
- Bao W, Yihong D, Lin Z, Zhu H. Free boundary models for mosquito range movement driven by climate warming. J Math Biol. 2018;76(4):841–875. - PubMed
-
- Barreira R, Elliott CM, Madzvamuse A. The surface finite element method for pattern formation on evolving biological surfaces. J Math Biol. 2011;63(6):1095–1119. - PubMed
-
- Batchelor GK (1967) An introduction to fluid dynamics. CUP
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

