Triggered Functional Dynamics of AsLOV2 by Time-Resolved Electron Paramagnetic Resonance at High Magnetic Fields
- PMID: 36638360
- PMCID: PMC12459748
- DOI: 10.1002/anie.202212832
Triggered Functional Dynamics of AsLOV2 by Time-Resolved Electron Paramagnetic Resonance at High Magnetic Fields
Abstract
We present time-resolved Gd-Gd electron paramagnetic resonance (TiGGER) at 240 GHz for tracking inter-residue distances during a protein's mechanical cycle in the solution state. TiGGER makes use of Gd-sTPATCN spin labels, whose favorable qualities include a spin-7/2 EPR-active center, short linker, narrow intrinsic linewidth, and virtually no anisotropy at high fields (8.6 T) when compared to nitroxide spin labels. Using TiGGER, we determined that upon light activation, the C-terminus and N-terminus of AsLOV2 separate in less than 1 s and relax back to equilibrium with a time constant of approximately 60 s. TiGGER revealed that the light-activated long-range mechanical motion is slowed in the Q513A variant of AsLOV2 and is correlated to the similarly slowed relaxation of the optically excited chromophore as described in recent literature. TiGGER has the potential to valuably complement existing methods for the study of triggered functional dynamics in proteins.
Keywords: EPR Spectroscopy; Gadolinium; Protein Structures; Tigger; Time-Resolved Spectroscopy.
© 2023 Wiley-VCH GmbH.
Figures

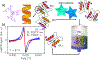


References
-
- Protein Data Bank, https://www.rcsb.org/ (visited on 06/15/2022).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

