Generation of anti-GD2 CAR macrophages from human pluripotent stem cells for cancer immunotherapies
- PMID: 36638788
- PMCID: PMC9968983
- DOI: 10.1016/j.stemcr.2022.12.012
Generation of anti-GD2 CAR macrophages from human pluripotent stem cells for cancer immunotherapies
Abstract
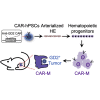
Macrophages armed with chimeric antigen receptors (CARs) provide a potent new option for treating solid tumors. However, genetic engineering and scalable production of somatic macrophages remains significant challenges. Here, we used CRISPR-Cas9 gene editing methods to integrate an anti-GD2 CAR into the AAVS1 locus of human pluripotent stem cells (hPSCs). We then established a serum- and feeder-free differentiation protocol for generating CAR macrophages (CAR-Ms) through arterial endothelial-to-hematopoietic transition (EHT). CAR-M produced by this method displayed a potent cytotoxic activity against GD2-expressing neuroblastoma and melanoma in vitro and neuroblastoma in vivo. This study provides a new platform for the efficient generation of off-the-shelf CAR-Ms for antitumor immunotherapy.
Keywords: CAR; GD2; PSCs; chimeric antigen receptor; hemogenic endothelium; immunotherapy; macrophages; melanoma; neuroblastoma; pluripotent stem cells.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interests C.M.C. receives honorarium for advisory board membership for Elephas Bio, Nektar Therapeutics, and Novartis. I.I.S serves on scientific advisory board for Umoja Biopharma. WARF has filed patent applications on the basis of this work, on which J.Z., A.M.., J.A.T., and I.I.S. are listed as inventors.
Figures



References
-
- Ahmed N., Brawley V., Hegde M., Bielamowicz K., Kalra M., Landi D., Robertson C., Gray T.L., Diouf O., Wakefield A., et al. HER2-Specific chimeric antigen receptor-modified virus-specific T cells for progressive glioblastoma: a phase 1 dose-escalation trial. JAMA Oncol. 2017;3:1094–1101. doi: 10.1001/jamaoncol.2017.0184. - DOI - PMC - PubMed
-
- Ahmed N., Brawley V.S., Hegde M., Robertson C., Ghazi A., Gerken C., Liu E., Dakhova O., Ashoori A., Corder A., et al. Human epidermal growth factor receptor 2 (HER2) -specific chimeric antigen receptor-modified T cells for the immunotherapy of HER2-positive sarcoma. J. Clin. Oncol. 2015;33:1688–1696. doi: 10.1200/JCO.2014.58.0225. - DOI - PMC - PubMed
-
- Boeva V., Louis-Brennetot C., Peltier A., Durand S., Pierre-Eugène C., Raynal V., Etchevers H.C., Thomas S., Lermine A., Daudigeos-Dubus E., et al. Heterogeneity of neuroblastoma cell identity defined by transcriptional circuitries. Nat. Genet. 2017;49:1408–1413. doi: 10.1038/ng.3921. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

