Control of protein stability by post-translational modifications
- PMID: 36639369
- PMCID: PMC9839724
- DOI: 10.1038/s41467-023-35795-8
Control of protein stability by post-translational modifications
Abstract
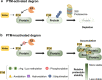
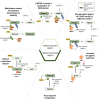
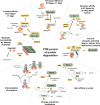
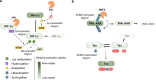
Post-translational modifications (PTMs) can occur on specific amino acids localized within regulatory domains of target proteins, which control a protein's stability. These regions, called degrons, are often controlled by PTMs, which act as signals to expedite protein degradation (PTM-activated degrons) or to forestall degradation and stabilize a protein (PTM-inactivated degrons). We summarize current knowledge of the regulation of protein stability by various PTMs. We aim to display the variety and breadth of known mechanisms of regulation as well as highlight common themes in PTM-regulated degrons to enhance potential for identifying novel drug targets where druggable targets are currently lacking.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

