Protein structure prediction has reached the single-structure frontier
- PMID: 36639584
- PMCID: PMC9839224
- DOI: 10.1038/s41592-022-01760-4
Protein structure prediction has reached the single-structure frontier
Abstract
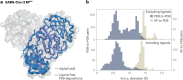
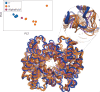
Dramatic advances in protein structure prediction have sparked debate as to whether the problem of predicting structure from sequence is solved or not. Here, I argue that AlphaFold2 and its peers are currently limited by the fact that they predict only a single structure, instead of a structural distribution, and that this realization is crucial for the next generation of structure prediction algorithms.
Conflict of interest statement
After submission but before final acceptance, the author became an employee and shareholder of CHARM Therapeutics Inc.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

