Neuronopathic GBA1L444P Mutation Accelerates Glucosylsphingosine Levels and Formation of Hippocampal Alpha-Synuclein Inclusions
- PMID: 36639889
- PMCID: PMC9864632
- DOI: 10.1523/JNEUROSCI.0680-22.2022
Neuronopathic GBA1L444P Mutation Accelerates Glucosylsphingosine Levels and Formation of Hippocampal Alpha-Synuclein Inclusions
Abstract
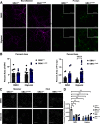
The most common genetic risk factor for Parkinson's disease (PD) is heterozygous mutations GBA1, which encodes for the lysosomal enzyme, glucocerebrosidase. Reduced glucocerebrosidase activity associates with an accumulation of abnormal α-synuclein (α-syn) called Lewy pathology, which characterizes PD. PD patients heterozygous for the neuronotypic GBA1L444P mutation (GBA1+/L444P) have a 5.6-fold increased risk of cognitive impairments. In this study, we used GBA1+/L444P mice of either sex to determine its effects on lipid metabolism, expression of synaptic proteins, behavior, and α-syn inclusion formation. At 3 months of age, GBA1+/L444P mice demonstrated impaired contextual fear conditioning, and increased motor activity. Hippocampal levels of vGLUT1 were selectively reduced in GBA1+/L444P mice. We show, using mass spectrometry, that GBA1L444P expression increased levels of glucosylsphingosine, but not glucosylceramide, in the brains and serum of GBA1+/L444P mice. Templated induction of α-syn pathology in mice showed an increase in α-syn inclusion formation in the hippocampus of GBA1+/L444P mice compared with GBA1+/+ mice, but not in the cortex, or substantia nigra pars compacta. Pathologic α-syn reduced SNc dopamine neurons by 50% in both GBA1+/+ and GBA1+/L444P mice. Treatment with a GlcCer synthase inhibitor did not affect abundance of α-syn inclusions in the hippocampus or rescue dopamine neuron loss. Overall, these data suggest the importance of evaluating the contribution of elevated glucosylsphingosine to PD phenotypes. Further, our data suggest that expression of neuronotypic GBA1L444P may cause defects in the hippocampus, which may be a mechanism by which cognitive decline is more prevalent in individuals with GBA1-PD.SIGNIFICANCE STATEMENT Parkinson's disease (PD) and dementia with Lewy bodies (DLB) are both pathologically characterized by abnormal α-synuclein (α-syn). Mutant GBA1 is a risk factor for both PD and DLB. Our data show the expression of neuronotypic GBA1L444P impairs behaviors related to hippocampal function, reduces expression of a hippocampal excitatory synaptic protein, and that the hippocampus is more susceptible to α-syn inclusion formation. Further, our data strengthen support for the importance of evaluating the contribution of glucosylsphingosine to PD phenotypes. These outcomes suggest potential mechanisms by which GBA1L444P contributes to the cognitive symptoms clinically observed in PD and DLB. Our findings also highlight the importance of glucosylsphingosine as a relevant biomarker for future therapeutics.
Keywords: Parkinson's disease; glucocerebrosidase; glucosylceramide; glucosylsphingosine; venglustat; α-synuclein.
Copyright © 2023 the authors.
Figures






References
-
- Arrant AE, Roth JR, Boyle NR, Kashyap SN, Hoffmann MQ, Murchison CF, Ramos EM, Nana AL, Spina S, Grinberg LT, Miller BL, Seeley WW, Roberson ED (2019) Impaired β-glucocerebrosidase activity and processing in frontotemporal dementia due to progranulin mutations. Acta Neuropathol Commun 7:17. 10.1186/s40478-019-0872-6 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous
