Specification of human germ cell fate with enhanced progression capability supported by hindgut organoids
- PMID: 36640324
- PMCID: PMC7618081
- DOI: 10.1016/j.celrep.2022.111907
Specification of human germ cell fate with enhanced progression capability supported by hindgut organoids
Abstract
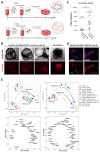
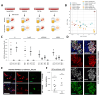
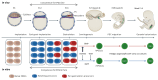
Human primordial germ cells (hPGCs), the precursors of sperm and eggs, are specified during weeks 2-3 after fertilization. Few studies on ex vivo and in vitro cultured human embryos reported plausible hPGCs on embryonic day (E) 12-13 and in an E16-17 gastrulating embryo. In vitro, hPGC-like cells (hPGCLCs) can be specified from the intermediary pluripotent stage or peri-gastrulation precursors. Here, we explore the broad spectrum of hPGCLC precursors and how different precursors impact hPGCLC development. We show that resetting precursors can give rise to hPGCLCs (rhPGCLCs) in response to BMP. Strikingly, rhPGCLCs co-cultured with human hindgut organoids progress at a pace reminiscent of in vivo hPGC development, unlike those derived from peri-gastrulation precursors. Moreover, rhPGCLC specification depends on both EOMES and TBXT, not just on EOMES as for peri-gastrulation hPGCLCs. Importantly, our study provides the foundation for developing efficient in vitro models of human gametogenesis.
Keywords: CP: Stem cell research; Hindgut organoid; Primordial germ cell; Primordial germ cell-like cell precursor.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




References
-
- Ohinata Y, Ohta H, Shigeta M, Yamanaka K, Wakayama T, Saitou M. A Signaling Principle for the Specification of the Germ Cell Lineage in Mice. Cell. 2009;137:571–584. - PubMed
-
- Ohinata Y, Payer B, O’Carroll D, Ancelin K, Ono Y, Sano M, Barton SC, Obukhanych T, Nussenzweig M, Tarakhovsky A, et al. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature. 2005;436:207–213. - PubMed
-
- Ying Y, Zhao G-Q. Cooperation of Endoderm-Derived BMP2 and Extraembryonic Ectoderm-Derived BMP4 in Primordial Germ Cell Generation in the Mouse. Developmental Biology. 2001;232:484–492. - PubMed
-
- Hayashi K, Ohta H, Kurimoto K, Aramaki S, Saitou M. Reconstitution of the Mouse Germ Cell Specification Pathway in Culture by Pluripotent Stem Cells. Cell. 2011;146:519–532. - PubMed
-
- Aramaki S, Hayashi K, Kurimoto K, Ohta H, Yabuta Y, Iwanari H, Mochizuki Y, Hamakubo T, Kato Y, Shirahige K, Saitou M. A Mesodermal Factor, T, Specifies Mouse Germ Cell Fate by Directly Activating Germline Determinants. Developmental Cell. 2013;27:516–529. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

