Spike-specific T cells are enriched in breastmilk following SARS-CoV-2 mRNA vaccination
- PMID: 36642379
- PMCID: PMC9836998
- DOI: 10.1016/j.mucimm.2023.01.003
Spike-specific T cells are enriched in breastmilk following SARS-CoV-2 mRNA vaccination
Abstract
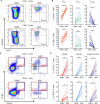
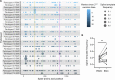
Human breastmilk is rich in T cells; however, their specificity and function are largely unknown. We compared the phenotype, diversity, and antigen specificity of T cells in breastmilk and peripheral blood of lactating individuals who received SARS-CoV-2 messenger RNA (mRNA) vaccination. Relative to blood, breastmilk contained higher frequencies of T effector and central memory populations that expressed mucosal-homing markers. T cell receptor sequence overlap was limited between blood and breastmilk. Overabundant breastmilk clones were observed in all individuals, were diverse, and contained complementarity-determining regions in three sequences with known epitope specificity, including to SARS-CoV-2 spike. SARS-CoV-2 spike-specific T cell receptors were more frequent in breastmilk compared to blood and expanded in breastmilk following a 3rd mRNA vaccine dose. Our observations indicate that the lactating breast contains a distinct T cell population that can be modulated by maternal vaccination with potential implications for passive infant protection.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures



Update of
-
Spike-specific T cells are enriched in breastmilk following SARS-CoV-2 mRNA vaccination.medRxiv [Preprint]. 2022 Sep 28:2021.12.03.21267036. doi: 10.1101/2021.12.03.21267036. medRxiv. 2022. Update in: Mucosal Immunol. 2023 Feb;16(1):39-49. doi: 10.1016/j.mucimm.2023.01.003. PMID: 36203549 Free PMC article. Updated. Preprint.
References
-
- Hassiotou F., Geddes D.T., Hartmann P.E. Cells in human milk: state of the science. J. Hum. Lact. 2013;29:171–182. - PubMed
-
- Goldman A.S., Garza C., Nichols B.L., Goldblum R.M. Immunologic factors in human milk during the first year of lactation. J. Pediatr. 1982;100:563–567. - PubMed
-
- Kent J.C. How breastfeeding works. J. Midwifery Womens Health. 2007;52:564–570. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

