AP-4 loss in CRISPR-edited zebrafish affects early embryo development
- PMID: 36642642
- PMCID: PMC9992121
- DOI: 10.1016/j.jbior.2022.100945
AP-4 loss in CRISPR-edited zebrafish affects early embryo development
Abstract
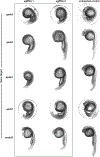
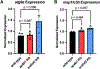
Mutations in the heterotetrametric adaptor protein 4 (AP-4; ε/β4/μ4/σ4 subunits) membrane trafficking coat complex lead to complex neurological disorders characterized by spastic paraplegia, microcephaly, and intellectual disabilities. Understanding molecular mechanisms underlying these disorders continues to emerge with recent identification of an essential autophagy protein, ATG9A, as an AP-4 cargo. Significant progress has been made uncovering AP-4 function in cell culture and patient-derived cell lines, and ATG9A trafficking by AP-4 is considered a potential target for gene therapy approaches. In contrast, understanding how AP-4 trafficking affects development and function at the organismal level has long been hindered by loss of conserved AP-4 genes in key model systems (S. cerevisiae, C. elegans, D. melanogaster). However, zebrafish (Danio rerio) have retained AP-4 and can serve as an important model system for studying both the nervous system and overall development. We undertook gene editing in zebrafish using a CRISPR-ExoCas9 knockout system to determine how loss of single AP-4, or its accessory protein tepsin, genes affect embryo development 24 h post-fertilization (hpf). Single gene-edited embryos display abnormal head morphology and neural necrosis. We further conducted the first exploration of how AP-4 single gene knockouts in zebrafish embryos affect expression levels and patterns of two autophagy genes, atg9a and map1lc3b. This work suggests zebrafish may be further adapted and developed as a tool to uncover AP-4 function in membrane trafficking and autophagy in the context of a model organism.
Keywords: CRISPR; Coat proteins; Gene editing; Membrane trafficking; Zebrafish.
Copyright © 2023 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare no competing conflicts of interest.
Figures


Similar articles
-
Adaptor protein complex 4 deficiency: a paradigm of childhood-onset hereditary spastic paraplegia caused by defective protein trafficking.Hum Mol Genet. 2020 Jan 15;29(2):320-334. doi: 10.1093/hmg/ddz310. Hum Mol Genet. 2020. PMID: 31915823 Free PMC article.
-
Bivalent Motif-Ear Interactions Mediate the Association of the Accessory Protein Tepsin with the AP-4 Adaptor Complex.J Biol Chem. 2015 Dec 25;290(52):30736-49. doi: 10.1074/jbc.M115.683409. Epub 2015 Nov 5. J Biol Chem. 2015. PMID: 26542808 Free PMC article.
-
Altered distribution of ATG9A and accumulation of axonal aggregates in neurons from a mouse model of AP-4 deficiency syndrome.PLoS Genet. 2018 Apr 26;14(4):e1007363. doi: 10.1371/journal.pgen.1007363. eCollection 2018 Apr. PLoS Genet. 2018. PMID: 29698489 Free PMC article.
-
The role of AP-4 in cargo export from the trans-Golgi network and hereditary spastic paraplegia.Biochem Soc Trans. 2020 Oct 30;48(5):1877-1888. doi: 10.1042/BST20190664. Biochem Soc Trans. 2020. PMID: 33084855 Review.
-
Adaptor protein complexes AP-4 and AP-5: new players in endosomal trafficking and progressive spastic paraplegia.Traffic. 2013 Feb;14(2):153-64. doi: 10.1111/tra.12028. Epub 2012 Dec 7. Traffic. 2013. PMID: 23167973 Review.
Cited by
-
Progress on multifunctional transmembrane protein ATG9A.Cell Commun Signal. 2025 Jul 1;23(1):314. doi: 10.1186/s12964-025-02317-6. Cell Commun Signal. 2025. PMID: 40598533 Free PMC article. Review.
-
Pluripotent Stem Cells as a Preclinical Cellular Model for Studying Hereditary Spastic Paraplegias.Int J Mol Sci. 2024 Feb 23;25(5):2615. doi: 10.3390/ijms25052615. Int J Mol Sci. 2024. PMID: 38473862 Free PMC article. Review.
References
-
- Abdollahpour H, Alawi M, Kortüm F, Beckstette M, Seemanova E, Komárek V, Rosenberger G, Kutsche K, 2014. An AP4B1 frameshift mutation in siblings with intellectual disability and spastic tetraplegia further delineates the AP-4 deficiency syndrome. European Journal of Human Genetics 2015 23:2 23, 256–259. - PMC - PubMed
-
- Abou Jamra R, Philippe O, Raas-Rothschild A, Eck SH, Graf E, Buchert R, Borck G, Ekici A, Brockschmidt FF, Nöthen MM, Munnich A, Strom TM, Reis A, Colleaux L, 2011. Adaptor protein complex 4 deficiency causes severe autosomal-recessive intellectual disability, progressive spastic paraplegia, shy character, and short stature. Am J Hum Genet 88, 788–795. - PMC - PubMed
-
- Bauer P, Leshinsky-Silver E, Blumkin L, Schlipf N, Schröder C, Schicks J, Lev D, Riess O, Lerman-Sagie T, Schöls L, 2012. Mutation in the AP4B1 gene cause hereditary spastic paraplegia type 47 (SPG47). Neurogenetics 13, 73–76. - PubMed
-
- Behne R, Teinert J, Wimmer M, D’Amore A, Davies AK, Scarrott JM, Eberhardt K, Brechmann B, Chen IPF, Buttermore ED, Barrett L, Dwyer S, Chen T, Hirst J, Wiesener A, Segal D, Martinuzzi A, Duarte ST, Bennett JT, Bourinaris T, Houlden H, Roubertie A, Santorelli FM, Robinson M, Azzouz M, Lipton JO, Borner GHH, Sahin M, Ebrahimi-Fakhari D, 2020. Adaptor protein complex 4 deficiency: a paradigm of childhood-onset hereditary spastic paraplegia caused by defective protein trafficking. Hum Mol Genet 29, 320–334. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

