The SmMYB36-SmERF6/SmERF115 module regulates the biosynthesis of tanshinones and phenolic acids in salvia miltiorrhiza hairy roots
- PMID: 36643739
- PMCID: PMC9832864
- DOI: 10.1093/hr/uhac238
The SmMYB36-SmERF6/SmERF115 module regulates the biosynthesis of tanshinones and phenolic acids in salvia miltiorrhiza hairy roots
Abstract
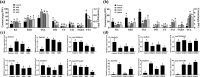
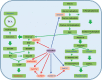
Tanshinone and phenolic acids are the most important active substances of Salvia miltiorrhiza, and the insight into their transcriptional regulatory mechanisms is an essential process to increase their content in vivo. SmMYB36 has been found to have important regulatory functions in the synthesis of tanshinone and phenolic acid; paradoxically, its mechanism of action in S. miltiorrhiza is not clear. Here, we demonstrated that SmMYB36 functions as a promoter of tanshinones accumulation and a suppressor of phenolic acids through the generation of SmMYB36 overexpressed and chimeric SmMYB36-SRDX (EAR repressive domain) repressor hairy roots in combination with transcriptomic-metabolomic analysis. SmMYB36 directly down-regulate the key enzyme gene of primary metabolism, SmGAPC, up-regulate the tanshinones biosynthesis branch genes SmDXS2, SmGGPPS1, SmCPS1 and down-regulate the phenolic acids biosynthesis branch enzyme gene, SmRAS. Meanwhile, SmERF6, a positive regulator of tanshinone synthesis activating SmCPS1, was up-regulated and SmERF115, a positive regulator of phenolic acid biosynthesis activating SmRAS, was down-regulated. Furthermore, the seven acidic amino acids at the C-terminus of SmMYB36 are required for both self-activating domain and activation of target gene expression. As a consequence, this study contributes to reveal the potential relevance of transcription factors synergistically regulating the biosynthesis of tanshinone and phenolic acid.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nanjing Agricultural University.
Figures


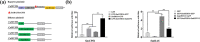


References
-
- Liu J, Osbourn A, Ma P. MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol Plant. 2015;8:689–708. - PubMed
-
- Ma D, Constabel CP. MYB repressors as regulators of phenylpropanoid metabolism in plants. Trends Plant Sci. 2019;24:275–89. - PubMed
-
- Tholl D. Biosynthesis and biological functions of terpenoids in plants. Adv Biochem Eng Biotechnol. 2015;148:63–106. - PubMed
-
- Mahjoub A, Hernould M, Joubès Jet al. . Overexpression of a grapevine R2R3-MYB factor in tomato affects vegetative development, flower morphology and flavonoid and terpenoid metabolism. Plant Physiol Biochem. 2009;47:551–61. - PubMed
-
- Zhu F, Luo T, Liu Cet al. . An R2R3-MYB transcription factor represses the transformation of α- and β-branch carotenoids by negatively regulating expression of CrBCH2 and CrNCED5 in flavedo of citrus reticulate. New Phytol. 2017;216:178–92. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

