Identification of novel antiviral drug candidates using an optimized SARS-CoV-2 phenotypic screening platform
- PMID: 36644320
- PMCID: PMC9822553
- DOI: 10.1016/j.isci.2023.105944
Identification of novel antiviral drug candidates using an optimized SARS-CoV-2 phenotypic screening platform
Abstract
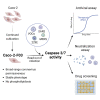
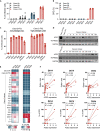
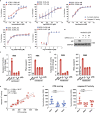
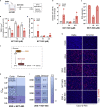
Reliable, easy-to-handle phenotypic screening platforms are needed for the identification of anti-SARS-CoV-2 compounds. Here, we present caspase 3/7 activity as a readout for monitoring the replication of SARS-CoV-2 isolates from different variants, including a remdesivir-resistant strain, and of other coronaviruses in numerous cell culture models, independently of cytopathogenic effect formation. Compared to other models, the Caco-2 subline Caco-2-F03 displayed superior performance. It possesses a stable SARS-CoV-2 susceptibility phenotype and does not produce false-positive hits due to drug-induced phospholipidosis. A proof-of-concept screen of 1,796 kinase inhibitors identified known and novel antiviral drug candidates including inhibitors of phosphoglycerate dehydrogenase (PHGDH), CDC like kinase 1 (CLK-1), and colony stimulating factor 1 receptor (CSF1R). The activity of the PHGDH inhibitor NCT-503 was further increased in combination with the hexokinase II (HK2) inhibitor 2-deoxy-D-glucose, which is in clinical development for COVID-19. In conclusion, caspase 3/7 activity detection in SARS-CoV-2-infected Caco-2-F03 cells provides a simple phenotypic high-throughput screening platform for SARS-CoV-2 drug candidates that reduces false-positive hits.
Keywords: Drugs; Screening in health technology; Virology.
© 2023 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Gao J., Sun F. Drug discovery to treat COVID-19 two years after its outbreak. Drug Discov. Ther. 2021;15:281–288. - PubMed
-
- Zhao H., Lu L., Peng Z., Chen L.L., Meng X., Zhang C., Ip J.D., Chan W.M., Chu A.W.H., Chan K.H., et al. SARS-CoV-2 Omicron variant shows less efficient replication and fusion activity when compared with Delta variant in TMPRSS2-expressed cells. Emerg. Microbes Infect. 2022;11:277–283. - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

