ScrepYard: An online resource for disulfide-stabilized tandem repeat peptides
- PMID: 36644825
- PMCID: PMC9885460
- DOI: 10.1002/pro.4566
ScrepYard: An online resource for disulfide-stabilized tandem repeat peptides
Abstract
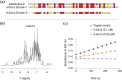
Receptor avidity through multivalency is a highly sought-after property of ligands. While readily available in nature in the form of bivalent antibodies, this property remains challenging to engineer in synthetic molecules. The discovery of several bivalent venom peptides containing two homologous and independently folded domains (in a tandem repeat arrangement) has provided a unique opportunity to better understand the underpinning design of multivalency in multimeric biomolecules, as well as how naturally occurring multivalent ligands can be identified. In previous work, we classified these molecules as a larger class termed secreted cysteine-rich repeat-proteins (SCREPs). Here, we present an online resource; ScrepYard, designed to assist researchers in identification of SCREP sequences of interest and to aid in characterizing this emerging class of biomolecules. Analysis of sequences within the ScrepYard reveals that two-domain tandem repeats constitute the most abundant SCREP domain architecture, while the interdomain "linker" regions connecting the functional domains are found to be abundant in amino acids with short or polar sidechains and contain an unusually high abundance of proline residues. Finally, we demonstrate the utility of ScrepYard as a virtual screening tool for discovery of putatively multivalent peptides, by using it as a resource to identify a previously uncharacterized serine protease inhibitor and confirm its predicted activity using an enzyme assay.
Keywords: SCREPs; bioactive; bivalent; disulfide-rich; multivalent; peptide; secreted proteins; tandem-repeat.
© 2023 The Authors. Protein Science published by Wiley Periodicals LLC on behalf of The Protein Society.
Conflict of interest statement
The authors have no conflicts of interest to declare.
Figures

References
-
- Armenteros JJA, Tsirigos KD, Sonderby CK, Petersen TN, Winther O, Brunak S, et al. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol. 2019;37:420–3. - PubMed
-
- Arner ES, Holmgren A. Physiological functions of thioredoxin and thioredoxin reductase. Eur J Biochem. 2000;267:6102–9. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

