Smartphone-read phage lateral flow assay for point-of-care detection of infection
- PMID: 36645184
- PMCID: PMC10503656
- DOI: 10.1039/d2an01499h
Smartphone-read phage lateral flow assay for point-of-care detection of infection
Erratum in
-
Correction: Smartphone-read phage lateral flow assay for point-of-care detection of infection.Analyst. 2024 Feb 26;149(5):1665. doi: 10.1039/d4an90016b. Analyst. 2024. PMID: 38348476 Free PMC article.
Abstract
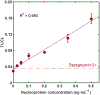
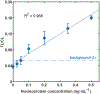
The COVID-19 pandemic has highlighted the urgent need for sensitive, affordable, and widely accessible testing at the point of care. Here we demonstrate a new, universal LFA platform technology using M13 phage conjugated with antibodies and HRP enzymes that offers high analytical sensitivity and excellent performance in a complex clinical matrix. We also report its complete integration into a sensitive chemiluminescence-based smartphone-readable lateral flow assay for the detection of SARS-CoV-2 nucleoprotein. We screened 84 anti-nucleoprotein monoclonal antibody pairs in phage LFA and identified an antibody pair that gave an LoD of 25 pg mL-1 nucleoprotein in nasal swab extract using a FluorChem gel documentation system and 100 pg mL-1 when the test was imaged and analyzed by an in-house-developed smartphone reader. The smartphone-read LFA signals for positive clinical samples tested (N = 15, with known Ct) were statistically different (p < 0.001) from signals for negative clinical samples (N = 11). The phage LFA technology combined with smartphone chemiluminescence imaging can enable the timely development of ultrasensitive, affordable point-of-care testing platforms for SARS-CoV-2 and beyond.
Conflict of interest statement
Conflicts of interest
Authors Binh Vu and Richard C. Willson are named inventors on IP which could relate to the subject of this paper.
Figures




References
-
- Frediani JK, Levy JM, Rao A, Bassit L, Figueroa J, Vos MB, Wood A, Jerris R, Leung-Pineda V, Gonzalez MD, Rogers BB, Mavigner M, Schinazi RF, Schoof N, Waggoner JJ, Kempker RR, Rebolledo PA, O’Neal JW, Stone C, Chahroudi A, Morris CR, Suessmith A, Sullivan J, Farmer S, Foster A, Roback JD, Ramachandra T, Washington C, Le K, Cordero MC, Esper A, Nehl EJ, Wang YF, Tyburski EA, Martin GS and Lam WA, Sci. Rep, 2021, 11, 14604. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

