Tryptophan Availability during Persistence of Chlamydia trachomatis Directly Impacts Expression of Chlamydial Cell Division Proteins
- PMID: 36645295
- PMCID: PMC9933654
- DOI: 10.1128/iai.00513-22
Tryptophan Availability during Persistence of Chlamydia trachomatis Directly Impacts Expression of Chlamydial Cell Division Proteins
Abstract
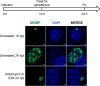
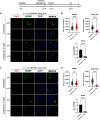
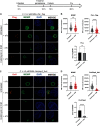
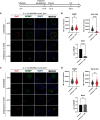
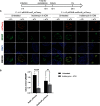
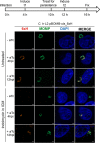
Chlamydia is an obligate intracellular pathogen with a highly reduced genome devoid of major stress response genes like relA and spoT, which mediate the stringent response. Interestingly, as an intracellular bacterium dependent on its host for nutrients and as a tryptophan (Trp) auxotroph, Chlamydia is very sensitive to Trp starvation, which is induced in vivo by the host cytokine interferon-γ. In response to Trp starvation, Chlamydia enters a viable but nonreplicating state called persistence. A major characteristic of chlamydial persistence is a block in cell division. We hypothesized that cell division is blocked during persistence by the inability to translate Trp-rich cell division proteins. To test this, we first investigated the translation of various cell division proteins under Trp starvation conditions using inducible expression strains. We observed that the Trp-poor protein MurG and the Trp-neutral protein FtsL were still expressed during persistence, while the expression of the Trp-rich proteins Pbp2, RodA, FtsI/Pbp3, and MraY was significantly reduced. As proof of concept for our hypothesis, we compared expression of a wild-type and mutant isoform of RodZ in which its four Trp codons were mutated. These experiments demonstrated that decreased expression of RodZ during persistence was reversed when no Trp was present in the protein, thus directly linking its expression to its Trp content. Together, these experiments indicate that specific cell division proteins are not produced during persistence. For the first time, our data provide a mechanism that explains the inhibition of cell division during chlamydial persistence mediated by Trp starvation.
Keywords: Chlamydia; cell division; peptidoglycan; persistence; persister; tryptophan.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Balaban NQ, Helaine S, Lewis K, Ackermann M, Aldridge B, Andersson DI, Brynildsen MP, Bumann D, Camilli A, Collins JJ, Dehio C, Fortune S, Ghigo J-M, Hardt W-D, Harms A, Heinemann M, Hung DT, Jenal U, Levin BR, Michiels J, Storz G, Tan M-W, Tenson T, Van Melderen L, Zinkernagel A. 2019. Definitions and guidelines for research on antibiotic persistence. Nat Rev Microbiol 17:441–448. 10.1038/s41579-019-0196-3. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

