Antiviral Approaches against Influenza Virus
- PMID: 36645300
- PMCID: PMC10035319
- DOI: 10.1128/cmr.00040-22
Antiviral Approaches against Influenza Virus
Abstract
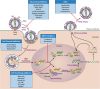
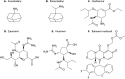
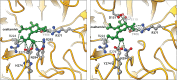
Preventing and controlling influenza virus infection remains a global public health challenge, as it causes seasonal epidemics to unexpected pandemics. These infections are responsible for high morbidity, mortality, and substantial economic impact. Vaccines are the prophylaxis mainstay in the fight against influenza. However, vaccination fails to confer complete protection due to inadequate vaccination coverages, vaccine shortages, and mismatches with circulating strains. Antivirals represent an important prophylactic and therapeutic measure to reduce influenza-associated morbidity and mortality, particularly in high-risk populations. Here, we review current FDA-approved influenza antivirals with their mechanisms of action, and different viral- and host-directed influenza antiviral approaches, including immunomodulatory interventions in clinical development. Furthermore, we also illustrate the potential utility of machine learning in developing next-generation antivirals against influenza.
Keywords: antiviral agents; drug resistance mechanisms; influenza; machine learning; monoclonal antibodies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

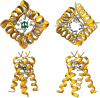
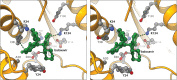
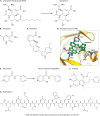
References
-
- World Health Organization. 2022. Comparison of number of influenza detections by subtype. https://app.powerbi.com/view?r=eyJrIjoiZTIxMzAwMzYtZWE4NC00YTU2LWE3MTUtM.... Retrieved 18 February 2022.
-
- Centers for Disease Control and Prevention. 2022. Weekly U.S. influenza surveillance report. https://www.cdc.gov/flu/weekly/index.htm. Retrieved 18 February 2022.
-
- Iuliano AD, Roguski KM, Chang HH, Muscatello DJ, Palekar R, Tempia S, Cohen C, Gran JM, Schanzer D, Cowling BJ, Wu P, Kyncl J, Ang LW, Park M, Redlberger-Fritz M, Yu H, Espenhain L, Krishnan A, Emukule G, van Asten L, Pereira da Silva S, Aungkulanon S, Buchholz U, Widdowson M-A, Bresee JS, Global Seasonal Influenza-associated Mortality Collaborator Network . 2018. Estimates of global seasonal influenza-associated respiratory mortality: a modelling study. Lancet 391:1285–1300. 10.1016/S0140-6736(17)33293-2. - DOI - PMC - PubMed
-
- Centers for Disease Control and Prevention. 2020. Past seasons estimated influenza disease burden. https://www.cdc.gov/flu/about/burden/past-seasons.html?web=1&wdLOR=cEAB9.... Retrieved 18 February 2022.
-
- Centers for Disease Control and Prevention. 2021. People at higher risk of flu complications, https://www.cdc.gov/flu/highrisk/index.htm. Retrieved 18 February 2022.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

