Determinant factors for residence time of kinesin motors at microtubule ends
- PMID: 36645568
- PMCID: PMC9958224
- DOI: 10.1007/s10867-022-09623-x
Determinant factors for residence time of kinesin motors at microtubule ends
Abstract
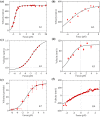
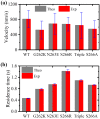
Kinesins constitute a superfamily of microtubule (MT)-based motor proteins, which can perform diverse biological functions in cells such as transporting vesicle, regulating MT dynamics, and segregating chromosome. Some motors such as kinesin-1, kinesin-2, and kinesin-3 do the activity mainly on the MT lattice, while others such as kinesin-7 and kinesin-8 do the activity mainly at the MT plus end. To perform the different functions, it is required that the former motors can reside on the MT lattice for longer times than at the end, while the latter motors can reside at the MT plus end for long times. Here, a simple but general theory of the MT-end residence time of the kinesin motor is presented, with which the factors dictating the residence time are determined. The theory is further used to study specifically the MT-end residence times of Drosophila kinesin-1, kinesin-2/KIF3AB, kinesin-3/Unc104, kinesin-5/Eg5, kinesin-7/CENP-E, and kinesin-8/Kip3 motors, with the theoretical results being in agreement with the available experimental data.
Keywords: Detachment time; Kinesin; Microtubule end; Molecular motor.
© 2023. The Author(s), under exclusive licence to Springer Nature B.V.
Conflict of interest statement
The author declares no competing interests.
Figures





Similar articles
-
Molecular mechanism of interaction between kinesin motors affecting their residence times on microtubule lattice and end.J Theor Biol. 2023 Aug 21;571:111556. doi: 10.1016/j.jtbi.2023.111556. Epub 2023 Jun 9. J Theor Biol. 2023. PMID: 37301429
-
A model of microtubule depolymerization by kinesin-8 motor proteins.Adv Protein Chem Struct Biol. 2024;141:87-122. doi: 10.1016/bs.apcsb.2023.12.002. Epub 2023 Dec 28. Adv Protein Chem Struct Biol. 2024. PMID: 38960488 Review.
-
Kinesin-14 motors participate in a force balance at microtubule plus-ends to regulate dynamic instability.Proc Natl Acad Sci U S A. 2022 Feb 22;119(8):e2108046119. doi: 10.1073/pnas.2108046119. Proc Natl Acad Sci U S A. 2022. PMID: 35173049 Free PMC article.
-
Biased Brownian motion as a mechanism to facilitate nanometer-scale exploration of the microtubule plus end by a kinesin-8.Proc Natl Acad Sci U S A. 2015 Jul 21;112(29):E3826-35. doi: 10.1073/pnas.1500272112. Epub 2015 Jul 6. Proc Natl Acad Sci U S A. 2015. PMID: 26150501 Free PMC article.
-
Microtubule-depolymerizing kinesins.Annu Rev Cell Dev Biol. 2013;29:417-41. doi: 10.1146/annurev-cellbio-101512-122345. Epub 2013 Jul 17. Annu Rev Cell Dev Biol. 2013. PMID: 23875646 Review.
Cited by
-
Modeling study of kinesin-13 MCAK microtubule depolymerase.Eur Biophys J. 2024 Aug;53(5-6):339-354. doi: 10.1007/s00249-024-01718-8. Epub 2024 Aug 2. Eur Biophys J. 2024. PMID: 39093405
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

