Cloning and expression study of a high-affinity nitrate transporter gene from Zea mays L
- PMID: 36645908
- PMCID: PMC9851203
- DOI: 10.1080/15592324.2022.2163342
Cloning and expression study of a high-affinity nitrate transporter gene from Zea mays L
Abstract
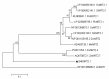
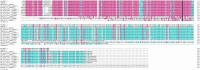
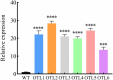
A nitrate transporter gene, named B46NRT2.1, from salt-tolerant Zea mays L. B46 has been cloned. B46NRT2.1 contained the same domain belonging to the major facilitator superfamily (PLN00028). The results of the phylogenetic tree indicated that B46NRT2.1 exhibits sequence similarity and the closest relationship with those known nitrate transporters of the NRT2 family. Through RT-qPCR, we found that the expression of B46NRT2.1 mainly happens in the root and leaf. Moreover, the treatment with NaCl, Na2CO3, and NaHCO3 could significantly increase the expression of B46NRT2.1. B46NRT2.1 was located in the plasma membrane. Through the study of yeast and plant salt response brought by B46NRT2.1 overexpression, we have preliminary knowledge that the expression of B46NRT2.1 makes yeast and plants respond to salt shock. There are 10 different kinds of cis-acting regulatory elements (CRES) in the promotor sequences of B46NRT2.1 gene using the PlantCARE web server to analyze. It mainly includes hormone response, abscisic acid, salicylic acid, gibberellin, methyl jasmonate, and auxin. The B46NRT2.1 gene's co-expression network showed that it was co-expressed with a number of other genes in several biological pathways, including regulation of NO3 long-distance transit, modulation of nitrate sensing and metabolism, nitrate assimilation, and transduction of Jasmonic acid-independent wound signal. The results of this work should serve as a good scientific foundation for further research on the functions of the NRT2 gene family in plants (inbred line B46), and this research adds to our understanding of the molecular mechanisms under salt tolerance.
Keywords: High-affinity nitrate transporter gene; Zea mays; salt shock.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures










Similar articles
-
[Identification, expression and DNA variation analysis of high affinity nitrate transporter NRT2/3 gene family in Sorghum bicolor].Sheng Wu Gong Cheng Xue Bao. 2023 Jul 25;39(7):2743-2761. doi: 10.13345/j.cjb.220800. Sheng Wu Gong Cheng Xue Bao. 2023. PMID: 37584129 Chinese.
-
Novel Proteins of the High-Affinity Nitrate Transporter Family NRT2, SaNRT2.1 and SaNRT2.5, from the Euhalophyte Suaeda altissima: Molecular Cloning and Expression Analysis.Int J Mol Sci. 2024 May 22;25(11):5648. doi: 10.3390/ijms25115648. Int J Mol Sci. 2024. PMID: 38891835 Free PMC article.
-
Genome-wide identification and characterization of high-affinity nitrate transporter 2 (NRT2) gene family in tomato (Solanum lycopersicum) and their transcriptional responses to drought and salinity stresses.J Plant Physiol. 2022 May;272:153684. doi: 10.1016/j.jplph.2022.153684. Epub 2022 Mar 23. J Plant Physiol. 2022. PMID: 35349936
-
Nitrate transporters: an overview in legumes.Planta. 2017 Oct;246(4):585-595. doi: 10.1007/s00425-017-2724-6. Epub 2017 Jun 26. Planta. 2017. PMID: 28653185 Review.
-
Functional and Molecular Characterization of Plant Nitrate Transporters Belonging to NPF (NRT1/PTR) 6 Subfamily.Int J Mol Sci. 2024 Dec 20;25(24):13648. doi: 10.3390/ijms252413648. Int J Mol Sci. 2024. PMID: 39769409 Free PMC article. Review.
Cited by
-
Genome-wide identification of high-affinity nitrate transporter 2 (NRT2) gene family under phytohormones and abiotic stresses in alfalfa (Medicago sativa).Sci Rep. 2024 Dec 30;14(1):31920. doi: 10.1038/s41598-024-83438-9. Sci Rep. 2024. PMID: 39738449 Free PMC article.
References
-
- Wu J, Jiang Y, Liang Y, Chen L, Chen W, Cheng B. Expression of the maize MYB transcription factor ZmMYB3R enhances drought and salt stress tolerance in transgenic plants. Plant Physiol Biochem. 2019;137:179–188. - PubMed
-
- Yu C, Yan M, Dong H, Luo J, Ke Y, Guo A, Chen Y, Zhang J, Huang X. Maize bHLH55 functions positively in salt tolerance through modulation of AsA biosynthesis by directly regulating GDP-mannose pathway genes. Plant Sci. 2021;302:110676. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
