Single-cell RNA-seq integrated with multi-omics reveals SERPINE2 as a target for metastasis in advanced renal cell carcinoma
- PMID: 36646679
- PMCID: PMC9842647
- DOI: 10.1038/s41419-023-05566-w
Single-cell RNA-seq integrated with multi-omics reveals SERPINE2 as a target for metastasis in advanced renal cell carcinoma
Abstract
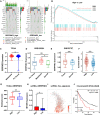
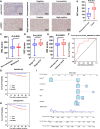
Tumor growth, metastasis and therapeutic response are believed to be regulated by the tumor and its microenvironment (TME) in advanced renal cell carcinoma (RCC). However, the mechanisms underlying genomic, transcriptomic and epigenetic alternations in RCC progression have not been completely defined. In this study, single-cell RNA-sequencing (scRNA-seq) data were obtained from eight tissue samples of RCC patients, including two matched pairs of primary and metastatic sites (lymph nodes), along with Hi-C, transposable accessible chromatin by high-throughput (ATAC-seq) and RNA-sequencing (RNA-seq) between RCC (Caki-1) and human renal tubular epithelial cell line (HK-2). The identified target was verified in clinical tissue samples (microarray of 407 RCC patients, TMA-30 and TMA-2020), whose function was further validated by in vitro and in vivo experiments through knockdown or overexpression. We profiled transcriptomes of 30514 malignant cells, and 14762 non-malignant cells. Comprehensive multi-omics analysis revealed that malignant cells and TME played a key role in RCC. The expression programs of stromal cells and immune cells were consistent among the samples, whereas malignant cells expressed distinct programs associated with hypoxia, cell cycle, epithelial differentiation, and two different metastasis patterns. Comparison of the hierarchical structure showed that SERPINE2 was related to these NNMF expression programs, and at the same time targeted the switched compartment. SERPINE2 was highly expressed in RCC tissues and lowly expressed in para-tumor tissues or HK-2 cell line. SERPINE2 knockdown markedly suppressed RCC cell growth and invasion, while SERPINE2 overexpression dramatically promoted RCC cell metastasis both in vitro and in vivo. In addition, SERPINE2 could activate the epithelial-mesenchymal transition pathway. The above findings demonstrated that the role of distinct expression patterns of malignant cells and TME played a distinct role in RCC progression. SERPINE2 was identified as a potential therapeutic target for inhibiting metastasis in advanced RCC.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

