Rapidly shifting immunologic landscape and severity of SARS-CoV-2 in the Omicron era in South Africa
- PMID: 36646700
- PMCID: PMC9842214
- DOI: 10.1038/s41467-022-35652-0
Rapidly shifting immunologic landscape and severity of SARS-CoV-2 in the Omicron era in South Africa
Abstract
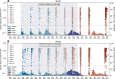
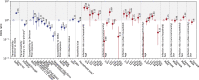
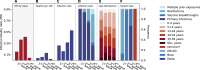
South Africa was among the first countries to detect the SARS-CoV-2 Omicron variant. However, the size of its Omicron BA.1 and BA.2 subvariants (BA.1/2) wave remains poorly understood. We analyzed sequential serum samples collected through a prospective cohort study before, during, and after the Omicron BA.1/2 wave to infer infection rates and monitor changes in the immune histories of participants over time. We found that the Omicron BA.1/2 wave infected more than half of the cohort population, with reinfections and vaccine breakthroughs accounting for > 60% of all infections in both rural and urban sites. After the Omicron BA.1/2 wave, we found few (< 6%) remained naïve to SARS-CoV-2 and the population immunologic landscape is fragmented with diverse infection/immunization histories. Prior infection with the ancestral strain, Beta, and Delta variants provided 13%, 34%, and 51% protection against Omicron BA.1/2 infection, respectively. Hybrid immunity and repeated prior infections reduced the risks of Omicron BA.1/2 infection by 60% and 85% respectively. Our study sheds light on a rapidly shifting landscape of population immunity in the Omicron era and provides context for anticipating the long-term circulation of SARS-CoV-2 in populations no longer naïve to the virus.
© 2023. The Author(s).
Conflict of interest statement
C.C. has received grant support from Sanofi Pasteur, Advanced Vaccine Initiative, and payment of travel costs from Parexel. Av.G. has received grant support from Sanofi Pasteur, Pfizer related to pneumococcal vaccine, C.D.C. and the Bill & Melinda Gates Foundation. N.W. reports grants from Sanofi Pasteur and the Bill & Melinda Gates Foundation. N.A.M. has received a grant to his institution from Pfizer to conduct research in patients with pneumonia and from Roche to collect specimens to assess a novel TB assay. J.M. has received grant support from Sanofi Pasteur. The remaining authors declare no competing interests.
Figures

Update of
-
Rapidly shifting immunologic landscape and severity of SARS-CoV-2 in the Omicron era in South Africa.medRxiv [Preprint]. 2022 Aug 22:2022.08.19.22278993. doi: 10.1101/2022.08.19.22278993. medRxiv. 2022. Update in: Nat Commun. 2023 Jan 16;14(1):246. doi: 10.1038/s41467-022-35652-0. PMID: 36032973 Free PMC article. Updated. Preprint.
References
-
- Tracking SARS-CoV-2 variants, (available at https://www.who.int/activities/tracking-SARS-CoV-2-variants).
-
- CoVariants, (available at https://covariants.org/).
-
- the Nextstrain team, Genomic epidemiology of novel coronavirus - Global subsampling. Nextstrainhttps://nextstrain.org/ncov/gisaid/global (2022).
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

