Expansion of interferon inducible gene pool via USP18 inhibition promotes cancer cell pyroptosis
- PMID: 36646704
- PMCID: PMC9842760
- DOI: 10.1038/s41467-022-35348-5
Expansion of interferon inducible gene pool via USP18 inhibition promotes cancer cell pyroptosis
Abstract
While immunotherapy has emerged as a breakthrough cancer therapy, it is only effective in some patients, indicating the need of alternative therapeutic strategies. Induction of cancer immunogenic cell death (ICD) is one promising way to elicit potent adaptive immune responses against tumor-associated antigens. Type I interferon (IFN) is well known to play important roles in different aspects of immune responses, including modulating ICD in anti-tumor action. However, how to expand IFN effect in promoting ICD responses has not been addressed. Here we show that depletion of ubiquitin specific protease 18 (USP18), a negative regulator of IFN signaling, selectively induces cancer cell ICD. Lower USP18 expression correlates with better survival across human selected cancer types and delays cancer progression in mouse models. Mechanistically, nuclear USP18 controls the enhancer landscape of cancer cells and diminishes STAT2-mediated transcription complex binding to IFN-responsive elements. Consequently, USP18 suppression not only enhances expression of canonical IFN-stimulated genes (ISGs), but also activates the expression of a set of atypical ISGs and NF-κB target genes, including genes such as Polo like kinase 2 (PLK2), that induce cancer pyroptosis. These findings may support the use of targeting USP18 as a potential cancer immunotherapy.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
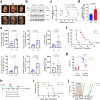
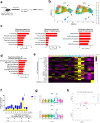
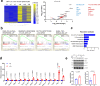






References
-
- Galluzzi L, Buque A, Kepp O, Zitvogel L, Kroemer G. Immunogenic cell death in cancer and infectious disease. Nat. Rev. Immunol. 2017;17:97–111. - PubMed
-
- Galon J, Bruni D. Approaches to treat immune hot, altered, and cold tumours with combination immunotherapies. Nat. Rev. Drug Discov. 2019;18:197–218. - PubMed
-
- Galluzzi L, Humeau J, Buque A, Zitvogel L, Kroemer G. Immunostimulation with chemotherapy in the era of immune checkpoint inhibitors. Nat. Rev. Clin. Oncol. 2020;17:725–741. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

