Rapid, multiplex detection of SARS-CoV-2 using isothermal amplification coupled with CRISPR-Cas12a
- PMID: 36646742
- PMCID: PMC9842216
- DOI: 10.1038/s41598-022-27133-7
Rapid, multiplex detection of SARS-CoV-2 using isothermal amplification coupled with CRISPR-Cas12a
Abstract
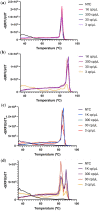
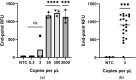
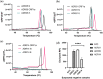
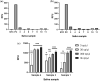
In December 2019 an outbreak erupted due to the beta coronavirus Severe Acute Respiratory Syndrome Coronavirus 2 in Wuhan, China. The disease caused by this virus (COVID-19) rapidly spread to all parts of the globe leading to a global pandemic. Efforts to combat the pandemic rely on RT-qPCR diagnostic tests that have high turnaround times (~ 24 h), are easily contaminated, need specialized equipment, facilities, and personnel that end up increasing the overall costs of this method. Loop-mediated isothermal amplification (LAMP) coupled with a reverse transcription step (RT-LAMP) is an alternative diagnostic method that can easily overcome these obstacles, when coupled with CRISPR/Cas it can eliminate false positives. Here we report a fast (~ 40 min), highly sensitive, point-of-care multiplex RT-LAMP and CRISPR/Cas12a assay to detect SARS-CoV-2. This fluorescence-based test achieved 100% specificity and 93% sensitivity using 25 positives and 50 negative patient samples for Ct < 35. Our reported LoD of 3 copies/µL will enable the robust, fast detection of the virus in a dedicated equipment which is a major step towards population-wide accessible testing.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
iSCAN: An RT-LAMP-coupled CRISPR-Cas12 module for rapid, sensitive detection of SARS-CoV-2.Virus Res. 2020 Oct 15;288:198129. doi: 10.1016/j.virusres.2020.198129. Epub 2020 Aug 18. Virus Res. 2020. PMID: 32822689 Free PMC article.
-
CLEVER assay: A visual and rapid RNA extraction-free detection of SARS-CoV-2 based on CRISPR-Cas integrated RT-LAMP technology.J Appl Microbiol. 2022 Aug;133(2):410-421. doi: 10.1111/jam.15571. Epub 2022 Apr 18. J Appl Microbiol. 2022. PMID: 35396760 Free PMC article.
-
Development and Validation of a Novel COVID-19 nsp8 One-Tube RT-LAMP-CRISPR Assay for SARS-CoV-2 Diagnosis.Microbiol Spectr. 2022 Dec 21;10(6):e0196222. doi: 10.1128/spectrum.01962-22. Epub 2022 Nov 29. Microbiol Spectr. 2022. PMID: 36445095 Free PMC article.
-
Diagnostic efficiency of RT-LAMP integrated CRISPR-Cas technique for COVID-19: A systematic review and meta-analysis.Pathog Glob Health. 2022 Oct;116(7):410-420. doi: 10.1080/20477724.2022.2035625. Epub 2022 Feb 10. Pathog Glob Health. 2022. PMID: 35142264 Free PMC article.
-
A Recent Update on Advanced Molecular Diagnostic Techniques for COVID-19 Pandemic: An Overview.Front Immunol. 2021 Dec 14;12:732756. doi: 10.3389/fimmu.2021.732756. eCollection 2021. Front Immunol. 2021. PMID: 34970254 Free PMC article. Review.
Cited by
-
Kinetic analysis and engineering of thermostable Cas12a for nucleic acid detection.Nucleic Acids Res. 2025 Jun 6;53(11):gkaf509. doi: 10.1093/nar/gkaf509. Nucleic Acids Res. 2025. PMID: 40512544 Free PMC article.
-
Laboratory-based molecular test alternatives to RT-PCR for the diagnosis of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2024 Oct 14;10(10):CD015618. doi: 10.1002/14651858.CD015618. Cochrane Database Syst Rev. 2024. PMID: 39400904
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

