Identification of biomarkers predictive of metastasis development in early-stage colorectal cancer using network-based regularization
- PMID: 36647008
- PMCID: PMC9841719
- DOI: 10.1186/s12859-022-05104-z
Identification of biomarkers predictive of metastasis development in early-stage colorectal cancer using network-based regularization
Abstract
Colorectal cancer (CRC) is the third most common cancer and the second most deathly worldwide. It is a very heterogeneous disease that can develop via distinct pathways where metastasis is the primary cause of death. Therefore, it is crucial to understand the molecular mechanisms underlying metastasis. RNA-sequencing is an essential tool used for studying the transcriptional landscape. However, the high-dimensionality of gene expression data makes selecting novel metastatic biomarkers problematic. To distinguish early-stage CRC patients at risk of developing metastasis from those that are not, three types of binary classification approaches were used: (1) classification methods (decision trees, linear and radial kernel support vector machines, logistic regression, and random forest) using differentially expressed genes (DEGs) as input features; (2) regularized logistic regression based on the Elastic Net penalty and the proposed iTwiner-a network-based regularizer accounting for gene correlation information; and (3) classification methods based on the genes pre-selected using regularized logistic regression. Classifiers using the DEGs as features showed similar results, with random forest showing the highest accuracy. Using regularized logistic regression on the full dataset yielded no improvement in the methods' accuracy. Further classification using the pre-selected genes found by different penalty factors, instead of the DEGs, significantly improved the accuracy of the binary classifiers. Moreover, the use of network-based correlation information (iTwiner) for gene selection produced the best classification results and the identification of more stable and robust gene sets. Some are known to be tumor suppressor genes (OPCML-IT2), to be related to resistance to cancer therapies (RAC1P3), or to be involved in several cancer processes such as genome stability (XRCC6P2), tumor growth and metastasis (MIR602) and regulation of gene transcription (NME2P2). We show that the classification of CRC patients based on pre-selected features by regularized logistic regression is a valuable alternative to using DEGs, significantly increasing the models' predictive performance. Moreover, the use of correlation-based penalization for biomarker selection stands as a promising strategy for predicting patients' groups based on RNA-seq data.
Keywords: Biomarker selection; Classification; Colorectal cancer; Regularization; iTwiner.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

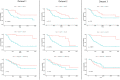
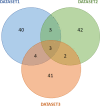
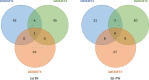
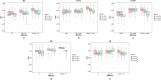
Similar articles
-
TCox: Correlation-Based Regularization Applied to Colorectal Cancer Survival Data.Biomedicines. 2020 Nov 10;8(11):488. doi: 10.3390/biomedicines8110488. Biomedicines. 2020. PMID: 33182598 Free PMC article.
-
A network-based predictive gene expression signature for recurrence risks in stage II colorectal cancer.Cancer Med. 2020 Jan;9(1):179-193. doi: 10.1002/cam4.2642. Epub 2019 Nov 14. Cancer Med. 2020. PMID: 31724326 Free PMC article.
-
Identifying the key genes and microRNAs in colorectal cancer liver metastasis by bioinformatics analysis and in vitro experiments.Oncol Rep. 2019 Jan;41(1):279-291. doi: 10.3892/or.2018.6840. Epub 2018 Nov 1. Oncol Rep. 2019. PMID: 30542696 Free PMC article.
-
MicroRNAs in colorectal cancer: role in metastasis and clinical perspectives.World J Gastroenterol. 2014 Dec 7;20(45):17011-9. doi: 10.3748/wjg.v20.i45.17011. World J Gastroenterol. 2014. PMID: 25493013 Free PMC article. Review.
-
New trends in molecular and cellular biomarker discovery for colorectal cancer.World J Gastroenterol. 2016 Jul 7;22(25):5678-93. doi: 10.3748/wjg.v22.i25.5678. World J Gastroenterol. 2016. PMID: 27433083 Free PMC article. Review.
Cited by
-
Development and validation of a biomarker-based prediction model for metastasis in patients with colorectal cancer: Application of machine learning algorithms.Heliyon. 2024 Dec 24;11(1):e41443. doi: 10.1016/j.heliyon.2024.e41443. eCollection 2025 Jan 15. Heliyon. 2024. PMID: 39839508 Free PMC article.
-
EsoDetect: computational validation and algorithm development of a novel diagnostic and prognostic tool for dysplasia in Barrett's esophagus.PeerJ. 2025 Jul 3;13:e19613. doi: 10.7717/peerj.19613. eCollection 2025. PeerJ. 2025. PMID: 40620772 Free PMC article.
-
Predicting patient outcomes with gene-expression biomarkers from colorectal cancer organoids and cell lines.Front Mol Biosci. 2025 Jan 15;12:1531175. doi: 10.3389/fmolb.2025.1531175. eCollection 2025. Front Mol Biosci. 2025. PMID: 39886381 Free PMC article.
-
Assessment of ID family proteins expression in colorectal cancer of Iraqi patients.Mol Biol Rep. 2024 Jul 13;51(1):806. doi: 10.1007/s11033-024-09775-0. Mol Biol Rep. 2024. PMID: 39001993
-
Prognostic value of SLC4A4 and its correlation with the microsatellite instability in colorectal cancer.Front Oncol. 2023 Apr 19;13:1179120. doi: 10.3389/fonc.2023.1179120. eCollection 2023. Front Oncol. 2023. PMID: 37152025 Free PMC article.
References
MeSH terms
Substances
Grants and funding
- 951970 (OLISSIPO project)/Horizon 2020
- 951970 (OLISSIPO project)/Horizon 2020
- PD/BD/139146/2018/Fundação para a Ciência e a Tecnologia
- CEECINST/00102/2018,UIDB/04516/2020 (NOVA LINCS), and UIDB/00297/2020 (CMA)/Fundação para a Ciência e a Tecnologia
- PIC/IC/82821/2007/Fundação para a Ciência e a Tecnologia
- PIC/IC/82821/2007/Fundação para a Ciência e a Tecnologia
- PIC/IC/82821/2007/Fundação para a Ciência e a Tecnologia
- PIC/IC/82821/2007/Fundação para a Ciência e a Tecnologia
- PIC/IC/82821/2007/Fundação para a Ciência e a Tecnologia
- PIC/IC/82821/2007/Fundação para a Ciência e a Tecnologia
- PIC/IC/82821/2007/Fundação para a Ciência e a Tecnologia
- PIC/IC/82821/2007/Fundação para a Ciência e a Tecnologia
- PIC/IC/82821/2007/Fundação para a Ciência e a Tecnologia
- PIC/IC/82821/2007/Fundação para a Ciência e a Tecnologia
- PIC/IC/82821/2007/Fundação para a Ciência e a Tecnologia
- PIC/IC/82821/2007/Fundação para a Ciência e a Tecnologia
- PIC/IC/82821/2007/Fundação para a Ciência e a Tecnologia
- PIC/IC/82821/2007/Fundação para a Ciência e a Tecnologia
- PIC/IC/82821/2007/Fundação para a Ciência e a Tecnologia
- PIC/IC/82821/2007/Fundação para a Ciência e a Tecnologia
- MONET (PTDC/CCI-BIO/4180/2020) and MATISSE (DSAIPA/DS/0026/2019)/Fundação para a Ciência e a Tecnologia
LinkOut - more resources
Full Text Sources
Medical

