Harmonising electronic health records for reproducible research: challenges, solutions and recommendations from a UK-wide COVID-19 research collaboration
- PMID: 36647111
- PMCID: PMC9842203
- DOI: 10.1186/s12911-022-02093-0
Harmonising electronic health records for reproducible research: challenges, solutions and recommendations from a UK-wide COVID-19 research collaboration
Abstract
Background: The CVD-COVID-UK consortium was formed to understand the relationship between COVID-19 and cardiovascular diseases through analyses of harmonised electronic health records (EHRs) across the four UK nations. Beyond COVID-19, data harmonisation and common approaches enable analysis within and across independent Trusted Research Environments. Here we describe the reproducible harmonisation method developed using large-scale EHRs in Wales to accommodate the fast and efficient implementation of cross-nation analysis in England and Wales as part of the CVD-COVID-UK programme. We characterise current challenges and share lessons learnt.
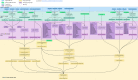
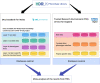
Methods: Serving the scope and scalability of multiple study protocols, we used linked, anonymised individual-level EHR, demographic and administrative data held within the SAIL Databank for the population of Wales. The harmonisation method was implemented as a four-layer reproducible process, starting from raw data in the first layer. Then each of the layers two to four is framed by, but not limited to, the characterised challenges and lessons learnt. We achieved curated data as part of our second layer, followed by extracting phenotyped data in the third layer. We captured any project-specific requirements in the fourth layer.
Results: Using the implemented four-layer harmonisation method, we retrieved approximately 100 health-related variables for the 3.2 million individuals in Wales, which are harmonised with corresponding variables for > 56 million individuals in England. We processed 13 data sources into the first layer of our harmonisation method: five of these are updated daily or weekly, and the rest at various frequencies providing sufficient data flow updates for frequent capturing of up-to-date demographic, administrative and clinical information.
Conclusions: We implemented an efficient, transparent, scalable, and reproducible harmonisation method that enables multi-nation collaborative research. With a current focus on COVID-19 and its relationship with cardiovascular outcomes, the harmonised data has supported a wide range of research activities across the UK.
Keywords: COVID-19; Common data model; Data harmonisation; Electronic health record; NHS digital TRE for England; Population health; Reproducible research; SAIL databank; Trusted Research Environments.
© 2023. The Author(s).
Conflict of interest statement
Not applicable.
Figures


References
-
- Health Dara Research UK. British Heart Foundation Data Science Centre [Internet]. 2022 [cited 2022 Aug 9]. Available from: https://www.hdruk.ac.uk/helping-with-health-data/bhf-data-science-centre/
-
- Vuokko R, Vakkuri A, Palojoki S. Harmonization of Finnish Vaccination Data. 2021 - PubMed
-
- Riffe T, Acosta E, Acosta EJ, Aburto DM, Alburez-Gutierrez A, Altová A, et al. Data resource profile: COVerAGE-DB: a global demographic database of COVID-19 cases and deaths. Int J Epidemiol. 2021;50(2):390–390f. doi: 10.1093/ije/dyab027. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

