Basal cell adhesion molecule promotes metastasis-associated processes in ovarian cancer
- PMID: 36647260
- PMCID: PMC9842900
- DOI: 10.1002/ctm2.1176
Basal cell adhesion molecule promotes metastasis-associated processes in ovarian cancer
Abstract
Background: Basal cell adhesion molecule (BCAM) is a laminin α5 (LAMA5) binding membrane-bound protein with a putative role in cancer. Besides full-length BCAM1, an isoform lacking most of the cytoplasmic domain (BCAM2), and a soluble form (sBCAM) of unknown function are known. In ovarian carcinoma (OC), all BCAM forms are abundant and associated with poor survival, yet BCAM's contribution to peritoneal metastatic spread remains enigmatic.
Methods: Biochemical, omics-based and real-time cell assays were employed to identify the source of sBCAM and metastasis-related functions of different BCAM forms. OC cells, explanted omentum and a mouse model of peritoneal colonisation were used in loss- and gain-of-function experiments.
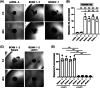
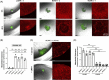
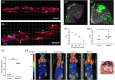
Results: We identified ADAM10 as a major BCAM sheddase produced by OC cells and identified proteolytic cleavage sites proximal to the transmembrane domain. Recombinant soluble BCAM inhibited single-cell adhesion and migration identically to membrane-bound isoforms, confirming its biological activity in OC. Intriguingly, this seemingly anti-tumorigenic potential of BCAM contrasts with a novel pro-metastatic function discovered in the present study. Thus, all queried BCAM forms decreased the compactness of tumour cell spheroids by inhibiting LAMA5 - integrin β1 interactions, promoted spheroid dispersion in a three-dimensional collagen matrix, induced clearance of mesothelial cells at spheroid attachment sites in vitro and enhanced invasion of spheroids into omental tissue both ex vivo and in vivo.
Conclusions: Membrane-bound BCAM as well as sBCAM shed by ADAM10 act as decoys rather than signalling receptors to modulate metastasis-related functions. While BCAM appears to have tumour-suppressive effects on single cells, it promotes the dispersion of OC cell spheroids by regulating LAMA5-integrin-β1-dependent compaction and thereby facilitating invasion of metastatic target sites. As peritoneal dissemination is majorly mediated by spheroids, these findings offer an explanation for the association of BCAM with a poor clinical outcome of OC, suggesting novel therapeutic options.
Keywords: ADAM10; BCAM; ovarian cancer; spheroids.
© 2023 The Authors. Clinical and Translational Medicine published by John Wiley & Sons Australia, Ltd on behalf of Shanghai Institute of Clinical Bioinformatics.
Conflict of interest statement
The authors declare no competing financial interests.
Figures





Similar articles
-
Basal cell adhesion molecule (BCAM) promotes mesothelial-to-mesenchymal transition and tumor angiogenesis through paracrine signaling.Cell Commun Signal. 2025 Mar 13;23(1):136. doi: 10.1186/s12964-025-02128-9. Cell Commun Signal. 2025. PMID: 40082910 Free PMC article.
-
BCAM and LAMA5 Mediate the Recognition between Tumor Cells and the Endothelium in the Metastatic Spreading of KRAS-Mutant Colorectal Cancer.Clin Cancer Res. 2016 Oct 1;22(19):4923-4933. doi: 10.1158/1078-0432.CCR-15-2664. Epub 2016 May 3. Clin Cancer Res. 2016. PMID: 27143691
-
Glycophorin-C sialylation regulates Lu/BCAM adhesive capacity during erythrocyte aging.Blood Adv. 2018 Jan 3;2(1):14-24. doi: 10.1182/bloodadvances.2017013094. eCollection 2018 Jan 9. Blood Adv. 2018. PMID: 29344581 Free PMC article.
-
Beyond tumor‑associated macrophages involved in spheroid formation and dissemination: Novel insights for ovarian cancer therapy (Review).Int J Oncol. 2024 Dec;65(6):117. doi: 10.3892/ijo.2024.5705. Epub 2024 Nov 8. Int J Oncol. 2024. PMID: 39513610 Free PMC article. Review.
-
Molecular and cellular mechanisms controlling integrin-mediated cell adhesion and tumor progression in ovarian cancer metastasis: a review.Clin Exp Metastasis. 2022 Apr;39(2):291-301. doi: 10.1007/s10585-021-10136-5. Epub 2021 Nov 25. Clin Exp Metastasis. 2022. PMID: 34822024 Free PMC article. Review.
Cited by
-
Stemness-driven clusters in ovarian cancer: immune characteristics and prognostic implications.Front Oncol. 2025 Jun 11;15:1577283. doi: 10.3389/fonc.2025.1577283. eCollection 2025. Front Oncol. 2025. PMID: 40567611 Free PMC article.
-
The Predictive Value of Neutrophil Extracellular Trap-Related Risk Score in Prognosis and Immune Microenvironment of Colorectal Cancer Patients.Mol Biotechnol. 2025 Apr;67(4):1509-1525. doi: 10.1007/s12033-024-01135-4. Epub 2024 Apr 5. Mol Biotechnol. 2025. PMID: 38580851
-
Basal cell adhesion molecule (BCAM) promotes mesothelial-to-mesenchymal transition and tumor angiogenesis through paracrine signaling.Cell Commun Signal. 2025 Mar 13;23(1):136. doi: 10.1186/s12964-025-02128-9. Cell Commun Signal. 2025. PMID: 40082910 Free PMC article.
-
BCAM (basal cell adhesion molecule) protein expression in different tumor populations.Discov Oncol. 2024 Aug 29;15(1):381. doi: 10.1007/s12672-024-01244-1. Discov Oncol. 2024. PMID: 39207605 Free PMC article.
-
Laminin α5: a key factor in tumor metastasis.Clin Exp Metastasis. 2025 Apr 11;42(3):24. doi: 10.1007/s10585-025-10341-6. Clin Exp Metastasis. 2025. PMID: 40214832 Review.
References
-
- Rahuel C, Le Van Kim C, Mattei MG, Cartron JP, Colin Y. A unique gene encodes spliceoforms of the B‐cell adhesion molecule cell surface glycoprotein of epithelial cancer and of the Lutheran blood group glycoprotein. Blood. 1996;88:1865‐1872. - PubMed
-
- Nemer El, W R, Colin Y, et al. Organization of the human LU gene and molecular basis of the Lu(a)/Lu(b) blood group polymorphism. Blood. 1997;89:4608‐4616. - PubMed
-
- Gauthier E, Nemer El, Wautier MP, et al. Role of the interaction between Lu/BCAM and the spectrin‐based membrane skeleton in the increased adhesion of hereditary spherocytosis red cells to laminin. Br J Haematol. 2010;148:456‐465. - PubMed
-
- Kikkawa Y, Sudo R, Kon J, et al. Laminin alpha 5 mediates ectopic adhesion of hepatocellular carcinoma through integrins and/or Lutheran/basal cell adhesion molecule. Exp Cell Res. 2008;314:2579‐2590. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
