Accurate Prediction of Enzyme Thermostabilization with Rosetta Using AlphaFold Ensembles
- PMID: 36647575
- PMCID: PMC9930118
- DOI: 10.1021/acs.jcim.2c01083
Accurate Prediction of Enzyme Thermostabilization with Rosetta Using AlphaFold Ensembles
Abstract
Thermostability enhancement is a fundamental aspect of protein engineering as a biocatalyst's half-life is key for its industrial and biotechnological application, particularly at high temperatures and under harsh conditions. Thermostability changes upon mutation originate from modifications of the free energy of unfolding (ΔGu), making thermostabilization extremely challenging to predict with computational methods. In this contribution, we combine global conformational sampling with energy prediction using AlphaFold and Rosetta to develop a new computational protocol for the quantitative prediction of thermostability changes upon laboratory evolution of acyltransferase LovD and lipase LipA. We highlight how using an ensemble of protein conformations rather than a single three-dimensional model is mandatory for accurate thermostability predictions. By comparing our approaches with existing ones, we show that ensembles based on AlphaFold models provide more accurate and robust calculated thermostability trends than ensembles based solely on crystallographic structures as the latter introduce a strong distortion (scaffold bias) in computed thermostabilities. Eliminating this bias is critical for computer-guided enzyme design and evaluating the effect of multiple mutations on protein stability.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
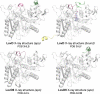
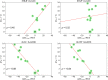
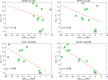
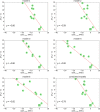

Similar articles
-
Rosetta Energy Analysis of AlphaFold2 models: Point Mutations and Conformational Ensembles.bioRxiv [Preprint]. 2024 Jan 27:2023.09.05.556364. doi: 10.1101/2023.09.05.556364. bioRxiv. 2024. PMID: 37732281 Free PMC article. Preprint.
-
Application of Rigidity Theory to the Thermostabilization of Lipase A from Bacillus subtilis.PLoS Comput Biol. 2016 Mar 22;12(3):e1004754. doi: 10.1371/journal.pcbi.1004754. eCollection 2016 Mar. PLoS Comput Biol. 2016. PMID: 27003415 Free PMC article.
-
Multidimensional computational strategies enhance the thermostability of alpha-galactosidase.Int J Biol Macromol. 2025 Jun;314:144316. doi: 10.1016/j.ijbiomac.2025.144316. Epub 2025 May 17. Int J Biol Macromol. 2025. PMID: 40388995
-
Recent advances in the improvement of enzyme thermostability by structure modification.Crit Rev Biotechnol. 2020 Feb;40(1):83-98. doi: 10.1080/07388551.2019.1682963. Epub 2019 Nov 5. Crit Rev Biotechnol. 2020. PMID: 31690132 Review.
-
Modeling conformational states of proteins with AlphaFold.Curr Opin Struct Biol. 2023 Aug;81:102645. doi: 10.1016/j.sbi.2023.102645. Epub 2023 Jun 29. Curr Opin Struct Biol. 2023. PMID: 37392556 Review.
Cited by
-
Structural Insights into Cold-Active Lipase from Glaciozyma antarctica PI12: Alphafold2 Prediction and Molecular Dynamics Simulation.J Mol Evol. 2024 Dec;92(6):944-963. doi: 10.1007/s00239-024-10219-3. Epub 2024 Nov 16. J Mol Evol. 2024. PMID: 39549052
-
Application of an alchemical free energy method for the prediction of thermostable DuraPETase variants.Appl Microbiol Biotechnol. 2024 Apr 21;108(1):305. doi: 10.1007/s00253-024-13144-z. Appl Microbiol Biotechnol. 2024. PMID: 38643427 Free PMC article.
-
Neural network conditioned to produce thermophilic protein sequences can increase thermal stability.Sci Rep. 2025 Apr 23;15(1):14124. doi: 10.1038/s41598-025-90828-0. Sci Rep. 2025. PMID: 40268970 Free PMC article.
-
Rosetta Energy Analysis of AlphaFold2 models: Point Mutations and Conformational Ensembles.bioRxiv [Preprint]. 2024 Jan 27:2023.09.05.556364. doi: 10.1101/2023.09.05.556364. bioRxiv. 2024. PMID: 37732281 Free PMC article. Preprint.
-
Rational design of lanosterol 14α-demethylase for ergosterol biosynthesis in Saccharomyces cerevisiae.3 Biotech. 2024 Dec;14(12):300. doi: 10.1007/s13205-024-04136-x. Epub 2024 Nov 15. 3 Biotech. 2024. PMID: 39554987
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

