Entry receptors - the gateway to alphavirus infection
- PMID: 36647825
- PMCID: PMC9843064
- DOI: 10.1172/JCI165307
Entry receptors - the gateway to alphavirus infection
Abstract
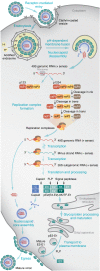
Alphaviruses are enveloped, insect-transmitted, positive-sense RNA viruses that infect humans and other animals and cause a range of clinical manifestations, including arthritis, musculoskeletal disease, meningitis, encephalitis, and death. Over the past four years, aided by CRISPR/Cas9-based genetic screening approaches, intensive research efforts have focused on identifying entry receptors for alphaviruses to better understand the basis for cellular and species tropism. Herein, we review approaches to alphavirus receptor identification and how these were used for discovery. The identification of new receptors advances our understanding of viral pathogenesis, tropism, and evolution and is expected to contribute to the development of novel strategies for prevention and treatment of alphavirus infection.
Conflict of interest statement
Figures


References
-
- Weaver SC, et al. Alphavirus infections. In: Guerrant RL, et al., eds. Tropical Infectious Diseases: Principles, Pathogens and Practice. 3rd ed. Saunders; 2011:519–524.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

