High-throughput telomere length measurement at nucleotide resolution using the PacBio high fidelity sequencing platform
- PMID: 36650155
- PMCID: PMC9845338
- DOI: 10.1038/s41467-023-35823-7
High-throughput telomere length measurement at nucleotide resolution using the PacBio high fidelity sequencing platform
Abstract
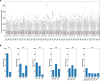
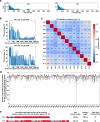
Telomeres are specialized nucleoprotein structures at the ends of linear chromosomes. The progressive shortening of steady-state telomere length in normal human somatic cells is a promising biomarker for age-associated diseases. However, there remain substantial challenges in quantifying telomere length due to the lack of high-throughput method with nucleotide resolution for individual telomere. Here, we describe a workflow to capture telomeres using newly designed telobaits in human culture cell lines as well as clinical patient samples and measure their length accurately at nucleotide resolution using single-molecule real-time (SMRT) sequencing. Our results also reveal the extreme heterogeneity of telomeric variant sequences (TVSs) that are dispersed throughout the telomere repeat region. The presence of TVSs disrupts the continuity of the canonical (5'-TTAGGG-3')n telomere repeats, which affects the binding of shelterin complexes at the chromosomal ends and telomere protection. These findings may have profound implications in human aging and diseases.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

