Rapid transmission and tight bottlenecks constrain the evolution of highly transmissible SARS-CoV-2 variants
- PMID: 36650162
- PMCID: PMC9844183
- DOI: 10.1038/s41467-023-36001-5
Rapid transmission and tight bottlenecks constrain the evolution of highly transmissible SARS-CoV-2 variants
Abstract
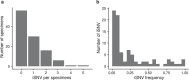
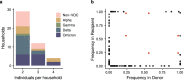
Transmission bottlenecks limit the spread of novel mutations and reduce the efficiency of selection along a transmission chain. While increased force of infection, receptor binding, or immune evasion may influence bottleneck size, the relationship between transmissibility and the transmission bottleneck is unclear. Here we compare the transmission bottleneck of non-VOC SARS-CoV-2 lineages to those of Alpha, Delta, and Omicron. We sequenced viruses from 168 individuals in 65 households. Most virus populations had 0-1 single nucleotide variants (iSNV). From 64 transmission pairs with detectable iSNV, we identify a per clade bottleneck of 1 (95% CI 1-1) for Alpha, Delta, and Omicron and 2 (95% CI 2-2) for non-VOC. These tight bottlenecks reflect the low diversity at the time of transmission, which may be more pronounced in rapidly transmissible variants. Tight bottlenecks will limit the development of highly mutated VOC in transmission chains, adding to the evidence that selection over prolonged infections may drive their evolution.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Update of
-
Rapid transmission and tight bottlenecks constrain the evolution of highly transmissible SARS-CoV-2 variants.bioRxiv [Preprint]. 2022 Oct 14:2022.10.12.511991. doi: 10.1101/2022.10.12.511991. bioRxiv. 2022. Update in: Nat Commun. 2023 Jan 17;14(1):272. doi: 10.1038/s41467-023-36001-5. PMID: 36263068 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

