SARS-CoV-2 Z-RNA activates the ZBP1-RIPK3 pathway to promote virus-induced inflammatory responses
- PMID: 36650286
- PMCID: PMC9844202
- DOI: 10.1038/s41422-022-00775-y
SARS-CoV-2 Z-RNA activates the ZBP1-RIPK3 pathway to promote virus-induced inflammatory responses
Abstract
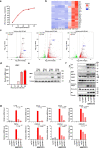
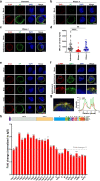
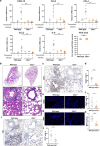
SARS-CoV-2 infection can trigger strong inflammatory responses and cause severe lung damage in COVID-19 patients with critical illness. However, the molecular mechanisms by which the infection induces excessive inflammatory responses are not fully understood. Here, we report that SARS-CoV-2 infection results in the formation of viral Z-RNA in the cytoplasm of infected cells and thereby activates the ZBP1-RIPK3 pathway. Pharmacological inhibition of RIPK3 by GSK872 or genetic deletion of MLKL reduced SARS-CoV-2-induced IL-1β release. ZBP1 or RIPK3 deficiency leads to reduced production of both inflammatory cytokines and chemokines during SARS-CoV-2 infection both in vitro and in vivo. Furthermore, deletion of ZBP1 or RIPK3 alleviated SARS-CoV-2 infection-induced immune cell infiltration and lung damage in infected mouse models. These results suggest that the ZBP1-RIPK3 pathway plays a critical role in SARS-CoV-2-induced inflammatory responses and lung damage. Our study provides novel insights into how SARS-CoV-2 infection triggers inflammatory responses and lung pathology, and implicates the therapeutic potential of targeting ZBP1-RIPK3 axis in treating COVID-19.
© 2023. The Author(s) under exclusive licence to Center for Excellence in Molecular Cell Science, Chinese Academy of Sciences.
Conflict of interest statement
The authors declare no competing interests.
Figures




Comment in
-
ZBP1 inflames the SARS-CoV-2-infected lung.Cell Res. 2023 May;33(5):333-334. doi: 10.1038/s41422-023-00784-5. Cell Res. 2023. PMID: 36792808 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

