Stromal niche inflammation mediated by IL-1 signalling is a targetable driver of haematopoietic ageing
- PMID: 36650381
- PMCID: PMC7614279
- DOI: 10.1038/s41556-022-01053-0
Stromal niche inflammation mediated by IL-1 signalling is a targetable driver of haematopoietic ageing
Abstract
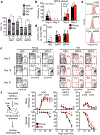
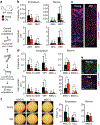
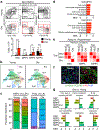
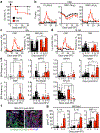
Haematopoietic ageing is marked by a loss of regenerative capacity and skewed differentiation from haematopoietic stem cells (HSCs), leading to impaired blood production. Signals from the bone marrow niche tailor blood production, but the contribution of the old niche to haematopoietic ageing remains unclear. Here we characterize the inflammatory milieu that drives both niche and haematopoietic remodelling. We find decreased numbers and functionality of osteoprogenitors at the endosteum and expansion of central marrow LepR+ mesenchymal stromal cells associated with deterioration of the sinusoidal vasculature. Together, they create a degraded and inflamed old bone marrow niche. Niche inflammation in turn drives the chronic activation of emergency myelopoiesis pathways in old HSCs and multipotent progenitors, which promotes myeloid differentiation and hinders haematopoietic regeneration. Moreover, we show how production of interleukin-1β (IL-1β) by the damaged endosteum acts in trans to drive the proinflammatory nature of the central marrow, with damaging consequences for the old blood system. Notably, niche deterioration, HSC dysfunction and defective regeneration can all be ameliorated by blocking IL-1 signalling. Our results demonstrate that targeting IL-1 as a key mediator of niche inflammation is a tractable strategy to improve blood production during ageing.
© 2023. The Author(s), under exclusive licence to Springer Nature Limited.
Figures










![Figure 7 ]](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/c1f4/10026258/756b6fba1561/nihms-1869586-f0007.gif)
References
-
- Rossi DJ, Jamieson CHM & Weissman IL Stems cells and the pathways to aging and cancer. Cell 132, 681–696 (2008). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

