Development of an assay system for the analysis of host RISC activity in the presence of a potyvirus RNA silencing suppressor, HC-Pro
- PMID: 36650505
- PMCID: PMC9844029
- DOI: 10.1186/s12985-022-01956-2
Development of an assay system for the analysis of host RISC activity in the presence of a potyvirus RNA silencing suppressor, HC-Pro
Abstract
Background: To investigate the mechanism of RNA silencing suppression, the genetic transformation of viral suppressors of RNA silencing (VSRs) in Arabidopsis integrates ectopic VSR expression at steady state, which overcomes the VSR variations caused by different virus infections or limitations of host range. Moreover, identifying the insertion of the transgenic VSR gene is necessary to establish a model transgenic plant for the functional study of VSR.
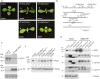
Methods: Developing an endogenous AGO1-based in vitro RNA-inducing silencing complex (RISC) assay prompts further investigation into VSR-mediated suppression. Three P1/HC-Pro plants from turnip mosaic virus (TuMV) (P1/HC-ProTu), zucchini yellow mosaic virus (ZYMV) (P1/HC-ProZy), and tobacco etch virus (TEV) (P1/HC-ProTe) were identified by T-DNA Finder and used as materials for investigations of the RISC cleavage efficiency.
Results: Our results indicated that the P1/HC-ProTu plant has slightly lower RISC activity than P1/HC-ProZy plants. In addition, the phenomena are consistent with those observed in TuMV-infected Arabidopsis plants, which implies that HC-ProTu could directly interfere with RISC activity.
Conclusions: In this study, we demonstrated the application of various plant materials in an in vitro RISC assay of VSR-mediated RNA silencing suppression.
Keywords: AGO1 degradation; HC-pro; In vitro RISC assay; T-DNA insertion.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources

