FASN multi-omic characterization reveals metabolic heterogeneity in pancreatic and prostate adenocarcinoma
- PMID: 36650542
- PMCID: PMC9847120
- DOI: 10.1186/s12967-023-03874-5
FASN multi-omic characterization reveals metabolic heterogeneity in pancreatic and prostate adenocarcinoma
Abstract
Background: Pancreatic ductal adenocarcinoma (PDAC) and prostate cancer (PCa) are among the most prevalent malignant tumors worldwide. There is now a comprehensive understanding of metabolic reprogramming as a hallmark of cancer. Fatty acid synthase (FASN) is a key regulator of the lipid metabolic network, providing energy to favor tumor proliferation and development. Whereas the biological role of FASN is known, its response and sensitivity to inhibition have not yet been fully established in these two cancer settings.
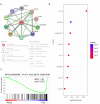
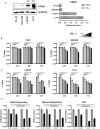
Methods: To evaluate the association between FASN expression, methylation, prognosis, and mutational profile in PDAC and PCa, we interrogated public databases and surveyed online platforms using TCGA data. The STRING database was used to investigate FASN interactors, and the Gene Set Enrichment Analysis platform Reactome database was used to perform an enrichment analysis using data from RNA sequencing public databases of PDAC and PCa. In vitro models using PDAC and PCa cell lines were used to corroborate the expression of FASN, as shown by Western blot, and the effects of FASN inhibition on cell proliferation/cell cycle progression and mitochondrial respiration were investigated with MTT, colony formation assay, cell cycle analysis and MitoStress Test.
Results: The expression of FASN was not modulated in PDAC compared to normal pancreatic tissues, while it was overexpressed in PCa, which also displayed a different level of promoter methylation. Based on tumor grade, FASN expression decreased in advanced stages of PDAC, but increased in PCa. A low incidence of FASN mutations was found for both tumors. FASN was overexpressed in PCa, despite not reaching statistical significance, and was associated with a worse prognosis than in PDAC. The biological role of FASN interactors correlated with lipid metabolism, and GSEA indicated that lipid-mediated mitochondrial respiration was enriched in PCa. Following validation of FASN overexpression in PCa compared to PDAC in vitro, we tested TVB-2640 as a FASN inhibitor. PCa proliferation arrest was modulated by FASN inhibition in a dose- and time-dependent manner, whereas PDAC proliferation was not altered. In line with this finding, mitochondrial respiration was found to be more affected in PCa than in PDAC. FASN inhibition interfered with metabolic signaling causing lipid accumulation and affecting cell viability with an impact on the replicative processes.
Conclusions: FASN exhibited differential expression patterns in PDAC and PCa, suggesting a different evolution during cancer progression. This was corroborated by the fact that both tumors responded differently to FASN inhibition in terms of proliferative potential and mitochondrial respiration, indicating that its use should reflect context specificity.
Keywords: FASN; Metabolism; Pancreatic adenocarcinoma; Proliferation; Prostate adenocarcinoma.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

