Phosphorylation of TRF2 promotes its interaction with TIN2 and regulates DNA damage response at telomeres
- PMID: 36651296
- PMCID: PMC9943673
- DOI: 10.1093/nar/gkac1269
Phosphorylation of TRF2 promotes its interaction with TIN2 and regulates DNA damage response at telomeres
Abstract
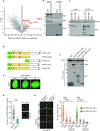
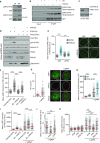
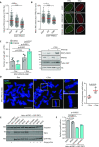
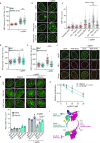
Protein phosphatase magnesium-dependent 1 delta (PPM1D) terminates the cell cycle checkpoint by dephosphorylating the tumour suppressor protein p53. By targeting additional substrates at chromatin, PPM1D contributes to the control of DNA damage response and DNA repair. Using proximity biotinylation followed by proteomic analysis, we identified a novel interaction between PPM1D and the shelterin complex that protects telomeric DNA. In addition, confocal microscopy revealed that endogenous PPM1D localises at telomeres. Further, we found that ATR phosphorylated TRF2 at S410 after induction of DNA double strand breaks at telomeres and this modification increased after inhibition or loss of PPM1D. TRF2 phosphorylation stimulated its interaction with TIN2 both in vitro and at telomeres. Conversely, induced expression of PPM1D impaired localisation of TIN2 and TPP1 at telomeres. Finally, recruitment of the DNA repair factor 53BP1 to the telomeric breaks was strongly reduced after inhibition of PPM1D and was rescued by the expression of TRF2-S410A mutant. Our results suggest that TRF2 phosphorylation promotes the association of TIN2 within the shelterin complex and regulates DNA repair at telomeres.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

