Identification of a Rho-Dependent Termination Site In Vivo Using Synthetic Small RNA
- PMID: 36651730
- PMCID: PMC9927376
- DOI: 10.1128/spectrum.03950-22
Identification of a Rho-Dependent Termination Site In Vivo Using Synthetic Small RNA
Abstract
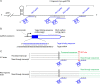
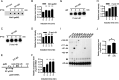
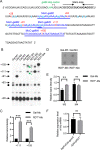
Rho promotes Rho-dependent termination (RDT) at the Rho-dependent terminator, producing a variable-length region without secondary structure at the 3' end of mRNA. Determining the exact RDT site in vivo is challenging, because the 3' end of mRNA is rapidly removed after RDT by 3'-to-5' exonuclease processing. Here, we applied synthetic small RNA (sysRNA) to identify the RDT region in vivo by exploiting its complementary base-pairing ability to target mRNA. Through the combined analyses of rapid amplification of cDNA 3' ends, primer extension, and capillary electrophoresis, we could precisely map and quantify mRNA 3' ends. We found that complementary double-stranded RNA (dsRNA) formed between sysRNA and mRNA was efficiently cleaved by RNase III in the middle of the dsRNA region. The formation of dsRNA appeared to protect the cleaved RNA 3' ends from rapid degradation by 3'-to-5' exonuclease, thereby stabilizing the mRNA 3' end. We further verified that the signal intensity at the 3' end was positively correlated with the amount of mRNA. By constructing a series of sysRNAs with close target sites and comparing the difference in signal intensity at the 3' end of wild-type and Rho-impaired strains, we finally identified a region of increased mRNA expression within the 21-bp range, which was determined as the RDT region. Our results demonstrated the ability to use sysRNA as a novel tool to identify RDT regions in vivo and expand the range of applications of sysRNA. IMPORTANCE sysRNA, which was formerly widely employed, has steadily lost popularity as more novel techniques for suppressing gene expression come into existence because of issues such as unstable inhibition effect and low inhibition efficiency. However, it remains an interesting topic as a regulatory tool due to its ease of design and low metabolic burden on cells. Here, for the first time, we discovered a new method to identify RDT regions in vivo using sysRNA. This new feature is important because since the discovery of the Rho protein in 1969, specific identification of RDT sites in vivo has been difficult due to the rapid processing of RNA 3' ends by exonucleases, and sysRNA might provide a new approach to address this challenge.
Keywords: RNA processing; RNase III; Rho-dependent termination; exonuclease; synthetic sRNA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

