The Klebsiella pneumoniae ter Operon Enhances Stress Tolerance
- PMID: 36651775
- PMCID: PMC9933665
- DOI: 10.1128/iai.00559-22
The Klebsiella pneumoniae ter Operon Enhances Stress Tolerance
Abstract
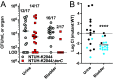
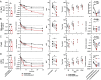
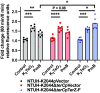
Healthcare-acquired infections are a leading cause of disease in patients that are hospitalized or in long-term-care facilities. Klebsiella pneumoniae (Kp) is a leading cause of bacteremia, pneumonia, and urinary tract infections in these settings. Previous studies have established that the ter operon, a genetic locus that confers tellurite oxide (K2TeO3) resistance, is associated with infection in colonized patients. Rather than enhancing fitness during infection, the ter operon increases Kp fitness during gut colonization; however, the biologically relevant function of this operon is unknown. First, using a murine model of urinary tract infection, we demonstrate a novel role for the ter operon protein TerC as a bladder fitness factor. To further characterize TerC, we explored a variety of functions, including resistance to metal-induced stress, resistance to radical oxygen species-induced stress, and growth on specific sugars, all of which were independent of TerC. Then, using well-defined experimental guidelines, we determined that TerC is necessary for tolerance to ofloxacin, polymyxin B, and cetylpyridinium chloride. We used an ordered transposon library constructed in a Kp strain lacking the ter operon to identify the genes that are required to resist K2TeO3-induced and polymyxin B-induced stress, which suggested that K2TeO3-induced stress is experienced at the bacterial cell envelope. Finally, we confirmed that K2TeO3 disrupts the Kp cell envelope, though these effects are independent of ter. Collectively, the results from these studies indicate a novel role for the ter operon as a stress tolerance factor, thereby explaining its role in enhancing fitness in the gut and bladder.
Keywords: Klebsiella; tolerance; transposons; urinary tract infection.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
The Klebsiella pneumoniae tellurium resistance gene terC contributes to both tellurite and zinc resistance.Microbiol Spectr. 2025 Apr 9;13(5):e0263424. doi: 10.1128/spectrum.02634-24. Online ahead of print. Microbiol Spectr. 2025. PMID: 40202338 Free PMC article.
-
A plasmid locus associated with Klebsiella clinical infections encodes a microbiome-dependent gut fitness factor.PLoS Pathog. 2021 Apr 30;17(4):e1009537. doi: 10.1371/journal.ppat.1009537. eCollection 2021 Apr. PLoS Pathog. 2021. PMID: 33930099 Free PMC article.
-
Klebsiella pneumoniae evolution in the gut leads to spontaneous capsule loss and decreased virulence potential.mBio. 2025 May 14;16(5):e0236224. doi: 10.1128/mbio.02362-24. Epub 2025 Mar 31. mBio. 2025. PMID: 40162782 Free PMC article.
-
Characterization of a novel chaperone/usher fimbrial operon present on KpGI-5, a methionine tRNA gene-associated genomic island in Klebsiella pneumoniae.BMC Microbiol. 2012 Apr 20;12:59. doi: 10.1186/1471-2180-12-59. BMC Microbiol. 2012. PMID: 22520965 Free PMC article.
-
Transcriptomic responses of a New Delhi metallo-β-lactamase-producing Klebsiella pneumoniae isolate to the combination of polymyxin B and chloramphenicol.Int J Antimicrob Agents. 2020 Aug;56(2):106061. doi: 10.1016/j.ijantimicag.2020.106061. Epub 2020 Jun 20. Int J Antimicrob Agents. 2020. PMID: 32574791
Cited by
-
TerC proteins function during protein secretion to metalate exoenzymes.Nat Commun. 2023 Oct 4;14(1):6186. doi: 10.1038/s41467-023-41896-1. Nat Commun. 2023. PMID: 37794032 Free PMC article.
-
Bacterial Metallostasis: Metal Sensing, Metalloproteome Remodeling, and Metal Trafficking.Chem Rev. 2024 Dec 25;124(24):13574-13659. doi: 10.1021/acs.chemrev.4c00264. Epub 2024 Dec 10. Chem Rev. 2024. PMID: 39658019 Free PMC article. Review.
-
The Klebsiella pneumoniae tellurium resistance gene terC contributes to both tellurite and zinc resistance.Microbiol Spectr. 2025 Apr 9;13(5):e0263424. doi: 10.1128/spectrum.02634-24. Online ahead of print. Microbiol Spectr. 2025. PMID: 40202338 Free PMC article.
-
Cross-border spread of a mosaic resistance (OXA-48) and virulence (aerobactin) plasmid in Klebsiella pneumoniae: a European Antimicrobial Resistance Genes Surveillance Network investigation, Europe, February 2019 to October 2024.Euro Surveill. 2025 Jul;30(27):2500439. doi: 10.2807/1560-7917.ES.2025.30.27.2500439. Euro Surveill. 2025. PMID: 40642770 Free PMC article.
-
Genomic Profile of a Multidrug-Resistant Klebsiella pneumoniae Strain Isolated from a Urine Specimen.Curr Microbiol. 2024 Jul 18;81(9):276. doi: 10.1007/s00284-024-03802-w. Curr Microbiol. 2024. PMID: 39023551
References
-
- Magill SS, Edwards JR, Bamberg W, Beldavs ZG, Dumyati G, Kainer MA, Lynfield R, Maloney M, McAllister-Hollod L, Nadle J, Ray SM, Thompson DL, Wilson LE, Fridkin SK, Multistate point-prevalence survey of health care–associated infections. 2014. N Engl J Med 370:1198–1208. doi:10.1056/NEJMoa1306801. - DOI - PMC - PubMed
-
- Wang M, Earley M, Chen L, Hanson BM, Yu Y, Liu Z, Salcedo S, Cober E, Li L, Kanj SS, Gao H, Munita JM, Ordoñez K, Weston G, Satlin MJ, Valderrama-Beltrán SL, Marimuthu K, Stryjewski ME, Komarow L, Luterbach C, Marshall SH, Rudin SD, Manca C, Paterson DL, Reyes J, Villegas MV, Evans S, Hill C, Arias R, Baum K, Fries BC, Doi Y, Patel R, Kreiswirth BN, Bonomo RA, Chambers HF, Fowler VG, Arias CA, van Duin D, Multi-Drug Resistant Organism Network I . 2022. Clinical outcomes and bacterial characteristics of carbapenem-resistant Klebsiella pneumoniae complex among patients from different global regions (CRACKLE-2): a prospective, multicentre, cohort study. Lancet Infect Dis 22:401–412. doi:10.1016/S1473-3099(21)00399-6. - DOI - PMC - PubMed
-
- Agyeman AA, Bergen PJ, Rao GG, Nation RL, Landersdorfer CB. 2020. A systematic review and meta-analysis of treatment outcomes following antibiotic therapy among patients with carbapenem-resistant Klebsiella pneumoniae infections. Int J Antimicrob Agents 55:105833. doi:10.1016/j.ijantimicag.2019.10.014. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

