A practical approach to curate clonal hematopoiesis of indeterminate potential in human genetic data sets
- PMID: 36652671
- PMCID: PMC10273159
- DOI: 10.1182/blood.2022018825
A practical approach to curate clonal hematopoiesis of indeterminate potential in human genetic data sets
Abstract
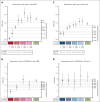
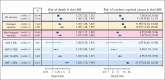
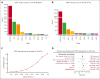
Clonal hematopoiesis of indeterminate potential (CHIP) is a common form of age-related somatic mosaicism that is associated with significant morbidity and mortality. CHIP mutations can be identified in peripheral blood samples that are sequenced using approaches that cover the whole genome, the whole exome, or targeted genetic regions; however, differentiating true CHIP mutations from sequencing artifacts and germ line variants is a considerable bioinformatic challenge. We present a stepwise method that combines filtering based on sequencing metrics, variant annotation, and population-based associations to increase the accuracy of CHIP calls. We apply this approach to ascertain CHIP in ∼550 000 individuals in the UK Biobank complete whole exome cohort and the All of Us Research Program initial whole genome release cohort. CHIP ascertainment on this scale unmasks recurrent artifactual variants and highlights the importance of specialized filtering approaches for several genes, including TET2 and ASXL1. We show how small changes in filtering parameters can considerably increase CHIP misclassification and reduce the effect size of epidemiological associations. Our high-fidelity call set refines previous population-based associations of CHIP with incident outcomes. For example, the annualized incidence of myeloid malignancy in individuals with small CHIP clones is 0.03% per year, which increases to 0.5% per year among individuals with very large CHIP clones. We also find a significantly lower prevalence of CHIP in individuals of self-reported Latino or Hispanic ethnicity in All of Us, highlighting the importance of including diverse populations. The standardization of CHIP calling will increase the fidelity of CHIP epidemiological work and is required for clinical CHIP diagnostic assays.
© 2023 by The American Society of Hematology.
Conflict of interest statement
Conflict-of-interest disclosure: M.S. has membership on a board or advisory committee of AbbVie, Bristol Myers Squibb, CTI, Forma, Geron, Karyopharm, Novartis, Ryvu, Sierra Oncology, Taiho, Takeda, and TG Therapeutics; has patents and royalties from Boehringer Ingelheim; received research funding from ALX Oncology, Astex, Incyte, Takeda, and TG Therapeutics; owns equity in Karyopharm and Ryvu; and is a consultant in: Forma, Karyopharm, and Ryvu. B.L.E. has received research funding from Celgene, Deerfield, and Novartis and consulting fees from GRAIL; and serves on the scientific advisory boards for Skyhawk Therapeutics, Exo Therapeutics, and Neomorph Therapeutics, all unrelated to this work. S.J. is a paid consultant to Novartis, AVRO Bio, Roche Genentech, GSK, and Foresite Labs and is on the scientific advisory board to Bitterroot Bio. B.L.E., S.J., A.B., and P.N. are cofounders, equity holders, and on the scientific advisory board of TenSixteen Bio. The remaining authors declare no competing financial interests.
Figures



Comment in
-
Refining CHIP in population data sets.Blood. 2023 May 4;141(18):2163-2164. doi: 10.1182/blood.2023019801. Blood. 2023. PMID: 37140953 No abstract available.
References
-
- Mustjoki S, Young NS. Somatic mutations in “Benign” disease. N Engl J Med. 2021;384(21):2039–2052. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

