Wnt/β-catenin signalling is required for pole-specific chromatin remodeling during planarian regeneration
- PMID: 36653403
- PMCID: PMC9849279
- DOI: 10.1038/s41467-023-35937-y
Wnt/β-catenin signalling is required for pole-specific chromatin remodeling during planarian regeneration
Abstract
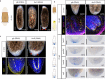
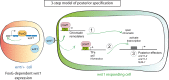
For successful regeneration, the identity of the missing tissue must be specified according to the pre-existing tissue. Planarians are ideal for the study of the mechanisms underlying this process; the same field of cells can regrow a head or a tail according to the missing body part. After amputation, the differential activation of the Wnt/β-catenin signal specifies anterior versus posterior identity. Initially, both wnt1 and notum (Wnt inhibitor) are expressed in all wounds, but 48 hours later they are restricted to posterior or anterior facing wounds, respectively, by an unknown mechanism. Here we show that 12 hours after amputation, the chromatin accessibility of cells in the wound region changes according to the polarity of the pre-existing tissue in a Wnt/β-catenin-dependent manner. Genomic analyses suggest that homeobox transcription factors and chromatin-remodeling proteins are direct Wnt/β-catenin targets, which trigger the expression of posterior effectors. Finally, we identify FoxG as a wnt1 up-stream regulator, probably via binding to its first intron enhancer region.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

