Genetic predictors of lifelong medication-use patterns in cardiometabolic diseases
- PMID: 36653479
- PMCID: PMC9873570
- DOI: 10.1038/s41591-022-02122-5
Genetic predictors of lifelong medication-use patterns in cardiometabolic diseases
Abstract
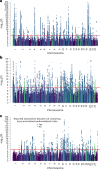
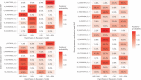
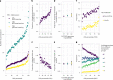
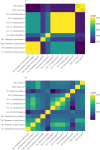
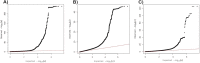
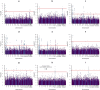
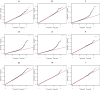
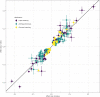
Little is known about the genetic determinants of medication use in preventing cardiometabolic diseases. Using the Finnish nationwide drug purchase registry with follow-up since 1995, we performed genome-wide association analyses of longitudinal patterns of medication use in hyperlipidemia, hypertension and type 2 diabetes in up to 193,933 individuals (55% women) in the FinnGen study. In meta-analyses of up to 567,671 individuals combining FinnGen with the Estonian Biobank and the UK Biobank, we discovered 333 independent loci (P < 5 × 10-9) associated with medication use. Fine-mapping revealed 494 95% credible sets associated with the total number of medication purchases, changes in medication combinations or treatment discontinuation, including 46 credible sets in 40 loci not associated with the underlying treatment targets. The polygenic risk scores (PRS) for cardiometabolic risk factors were strongly associated with the medication-use behavior. A medication-use enhanced multitrait PRS for coronary artery disease matched the performance of a risk factor-based multitrait coronary artery disease PRS in an independent sample (UK Biobank, n = 343,676). In summary, we demonstrate medication-based strategies for identifying cardiometabolic risk loci and provide genome-wide tools for preventing cardiovascular diseases.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Comment in
-
Genetic prediction of medication use patterns in cardiometabolic disease.Nat Med. 2023 Jan;29(1):43-44. doi: 10.1038/s41591-022-02124-3. Nat Med. 2023. PMID: 36683070 No abstract available.
References
-
- James SL, et al. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2018;392:1789–1858. doi: 10.1016/S0140-6736(18)32279-7. - DOI - PMC - PubMed
-
- Arnett DK, et al. 2019 ACC/AHA guideline on the primary prevention of cardiovascular disease: a report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. J. Am. Coll. Cardiol. 2019;74:e177–e232. doi: 10.1016/j.jacc.2019.03.010. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

