Structural Basis for Cyclosporin Isoform-Specific Inhibition of Cyclophilins from Toxoplasma gondii
- PMID: 36653744
- PMCID: PMC9926490
- DOI: 10.1021/acsinfecdis.2c00566
Structural Basis for Cyclosporin Isoform-Specific Inhibition of Cyclophilins from Toxoplasma gondii
Abstract
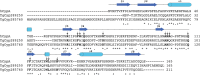
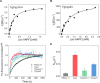
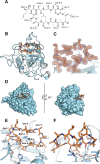
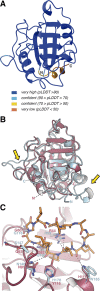
Cyclosporin (CsA) has antiparasite activity against the human pathogen Toxoplasma gondii. A possible mechanism of action involves CsA binding to T. gondii cyclophilins, although much remains to be understood. Herein, we characterize the functional and structural properties of a conserved (TgCyp23) and a more divergent (TgCyp18.4) cyclophilin isoform from T. gondii. While TgCyp23 is a highly active cis-trans-prolyl isomerase (PPIase) and binds CsA with nanomolar affinity, TgCyp18.4 shows low PPIase activity and is significantly less sensitive to CsA inhibition. The crystal structure of the TgCyp23:CsA complex was solved at the atomic resolution showing the molecular details of CsA recognition by the protein. Computational and structural studies revealed relevant differences at the CsA-binding site between TgCyp18.4 and TgCyp23, suggesting that the two cyclophilins might have distinct functions in the parasite. These studies highlight the extensive diversification of TgCyps and pave the way for antiparasite interventions based on selective targeting of cyclophilins.
Keywords: Toxoplasma gondii; chaperone-like activity; crystal structure; cyclophilin; cyclosporin; peptidyl-prolyl isomerization.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

