Chicken or Porcine Aminopeptidase N Mediates Cellular Entry of Pseudoviruses Carrying Spike Glycoprotein from the Avian Deltacoronaviruses HKU11, HKU13, and HKU17
- PMID: 36656013
- PMCID: PMC9973037
- DOI: 10.1128/jvi.01947-22
Chicken or Porcine Aminopeptidase N Mediates Cellular Entry of Pseudoviruses Carrying Spike Glycoprotein from the Avian Deltacoronaviruses HKU11, HKU13, and HKU17
Erratum in
-
Correction for Liang et al., "Chicken or Porcine Aminopeptidase N Mediates Cellular Entry of Pseudoviruses Carrying Spike Glycoprotein from the Avian Deltacoronaviruses HKU11, HKU13, and HKU17".J Virol. 2024 Dec 17;98(12):e0163124. doi: 10.1128/jvi.01631-24. Epub 2024 Nov 6. J Virol. 2024. PMID: 39503499 Free PMC article. No abstract available.
Abstract
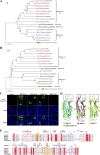
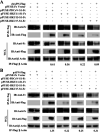
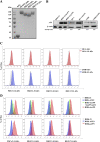
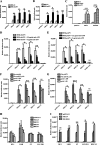
Members of deltacoronavirus (DCoV) have mostly been identified in diverse avian species as natural reservoirs, though the porcine DCoV (PDCoV) is a major swine enteropathogenic virus with global spread. The important role of aminopeptidase N (APN) orthologues from various mammalian and avian species in PDCoV cellular entry and interspecies transmission has been revealed recently. In this study, comparative analysis indicated that three avian DCoVs, bulbul DCoV HKU11, munia DCoV HKU13, and sparrow DCoV HKU17 (Chinese strain), and PDCoV in the subgenera Buldecovirus are grouped together at whole-genome levels; however, the spike (S) glycoprotein and its S1 subunit of HKU17 are more closely related to night heron DCoV HKU19 in Herdecovirus. Nevertheless, the S1 protein of HKU11, HKU13, or HKU17 bound to or interacted with chicken APN (chAPN) or porcine APN (pAPN) by flow cytometry analysis of cell surface expression of APN and by coimmunoprecipitation in APN-overexpressing cells. Expression of chAPN or pAPN allowed entry of pseudotyped lentiviruses with the S proteins from HKU11, HKU13 and HKU17 into nonsusceptible cells and natural avian and porcine cells, which could be inhibited by the antibody against APN or anti-PDCoV-S1. APN knockdown by siRNA or knockout by CRISPR/Cas9 in chicken or swine cell lines significantly or almost completely blocked infection of these pseudoviruses. Hence, we demonstrate that HKU11, HKU13, and HKU17 with divergent S genes likely engage chAPN or pAPN to enter the cells, suggesting a potential interspecies transmission from wild birds to poultry and from birds to mammals by certain avian DCoVs. IMPORTANCE The receptor usage of avian deltacoronaviruses (DCoVs) has not been investigated thus far, though porcine deltacoronavirus (PDCoV) has been shown to utilize aminopeptidase N (APN) as a cell receptor. We report here that chicken or porcine APN also mediates cellular entry by three avian DCoV (HKU11, HKU13, and HKU17) spike pseudoviruses, and the S1 subunit of three avian DCoVs binds to APN in vitro and in the surface of avian and porcine cells. The results fill the gaps in knowledge about the avian DCoV receptor and elucidate important insights for the monitoring and prevention of potential interspecies transmission of certain avian DCoVs. In view of the diversity of DCoVs, whether this coronavirus genus will cause novel virus to emerge in other mammals from birds, are worthy of further surveillance and investigation.
Keywords: aminopeptidase N (APN); avian deltacoronavirus; interspecies transmission; porcine deltacoronavirus (PDCoV); receptor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- de Groot RJ, Baker SC, Baric R, Enjuanes L, Gorbalenya AE, Holmes KV, Perlman S, Poon L, Rottier PJM, Talbot PJ, Woo PCY, Ziebuhr J. 2011. Coronaviridae, p 806–828. In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (ed), Virus taxonomy: ninth report of the international committee on taxonomy of viruses. Elsevier Academic Press, London, England.
-
- Woo PC, Lau SK, Lam CS, Lau CC, Tsang AK, Lau JH, Bai R, Teng JL, Tsang CC, Wang M, Zheng BJ, Chan KH, Yuen KY. 2012. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol 86:3995–4008. 10.1128/JVI.06540-11. - DOI - PMC - PubMed
-
- Lau SKP, Wong EYM, Tsang CC, Ahmed SS, Au-Yeung RKH, Yuen KY, Wernery U, Woo PCY. 2018. Discovery and sequence analysis of four deltacoronaviruses from birds in the middle east reveal interspecies jumping with recombination as a potential mechanism for avian-to-avian and avian-to-mammalian transmission. J Virol 92. 10.1128/JVI.00265-18. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

