A knockout-first model of H3f3a gene targeting leads to developmental lethality
- PMID: 36656301
- PMCID: PMC10038898
- DOI: 10.1002/dvg.23507
A knockout-first model of H3f3a gene targeting leads to developmental lethality
Abstract
Histone variant H3.3 is encoded by two genes, H3f3a and H3f3b, which can be expressed differentially depending on tissue type. Previous work in our lab has shown that knockout of H3f3b causes some neonatal lethality and infertility in mice, and chromosomal defects in mouse embryonic fibroblasts (MEFs). Studies of H3f3a and H3f3b null mice by others have produced generally similar phenotypes to what we found in our H3f3b nulls, but the relative impacts of the loss of either H3f3a or H3f3b have varied depending on the approach and genetic background. Here we used a knockout-first approach to target the H3f3a gene for inactivation in C57BL6 mice. Homozygous H3f3a targeting produced a lethal phenotype at or before birth. E13.5 null embryos had some potential morphological differences from WT littermates including smaller size and reduced head size. An E18.5 null embryo was smaller than its control littermates with several potential defects including small head and brain size as well as small lungs, which would be consistent with a late gestation lethal phenotype. Despite a reduction in H3.3 and total H3 protein levels, the only histone H3 post-translational modification in the small panel assessed that was significantly altered was the unique H3.3 mark phospho-Serine31, which was consistently increased in null neurospheres. H3f3a null neurospheres also exhibited consistent gene expression changes including in protocadherins. Overall, our findings are consistent with the model that there are differential, cell-type-specific contributions of H3f3a and H3f3b to H3.3 functions in epigenetic and developmental processes.
Keywords: H3f3a; H3f3b; development; histone H3.3; mouse knockouts; neurospheres.
© 2023 Wiley Periodicals LLC.
Figures
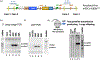



References
-
- Ahmad K, & Henikoff S (2002). The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Molecular Cell, 9(6), 1191–1200 http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dop... - PubMed
-
- Bryant L, Li D, Cox SG, Marchione D, Joiner EF, Wilson K, … Bhoj EJ (2020). Histone H3.3 beyond cancer: Germline mutations in histone 3 family 3A and 3B cause a previously unidentified neurodegenerative disorder in 46 patients. Science Advances, 6(49), eabc9207. 10.1126/sciadv.abc9207 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

