Gene Modulation with CRISPR-based Tools in Human iPSC-Cardiomyocytes
- PMID: 36656467
- PMCID: PMC9851124
- DOI: 10.1007/s12015-023-10506-4
Gene Modulation with CRISPR-based Tools in Human iPSC-Cardiomyocytes
Abstract
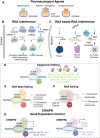
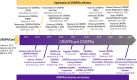
Precise control of gene expression (knock-out, knock-in, knockdown or overexpression) is at the heart of functional genomics - an approach to dissect the contribution of a gene/protein to the system's function. The development of a human in vitro system that can be patient-specific, induced pluripotent stem cells, iPSC, and the ability to obtain various cell types of interest, have empowered human disease modeling and therapeutic development. Scalable tools have been deployed for gene modulation in these cells and derivatives, including pharmacological means, DNA-based RNA interference and standard RNA interference (shRNA/siRNA). The CRISPR/Cas9 gene editing system, borrowed from bacteria and adopted for use in mammalian cells a decade ago, offers cell-specific genetic targeting and versatility. Outside genome editing, more subtle, time-resolved gene modulation is possible by using a catalytically "dead" Cas9 enzyme linked to an effector of gene transcription in combination with a guide RNA. The CRISPRi / CRISPRa (interference/activation) system evolved over the last decade as a scalable technology for performing functional genomics with libraries of gRNAs. Here, we review key developments of these approaches and their deployment in cardiovascular research. We discuss specific use with iPSC-cardiomyocytes and the challenges in further translation of these techniques.
Keywords: CRISPR; CRISPRa; CRISPRi; Gene knockdown; Gene modulation; Human iPSC-CMs; dCas9.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests or conflict of interest.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

