An atypical NLR protein modulates the NRC immune receptor network in Nicotiana benthamiana
- PMID: 36656829
- PMCID: PMC9851556
- DOI: 10.1371/journal.pgen.1010500
An atypical NLR protein modulates the NRC immune receptor network in Nicotiana benthamiana
Abstract
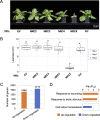
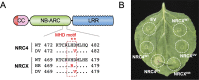
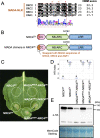
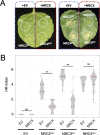
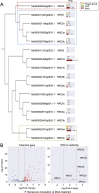
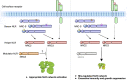
The NRC immune receptor network has evolved in asterid plants from a pair of linked genes into a genetically dispersed and phylogenetically structured network of sensor and helper NLR (nucleotide-binding domain and leucine-rich repeat-containing) proteins. In some species, such as the model plant Nicotiana benthamiana and other Solanaceae, the NRC (NLR-REQUIRED FOR CELL DEATH) network forms up to half of the NLRome, and NRCs are scattered throughout the genome in gene clusters of varying complexities. Here, we describe NRCX, an atypical member of the NRC family that lacks canonical features of these NLR helper proteins, such as a functional N-terminal MADA motif and the capacity to trigger autoimmunity. In contrast to other NRCs, systemic gene silencing of NRCX in N. benthamiana markedly impairs plant growth resulting in a dwarf phenotype. Remarkably, dwarfism of NRCX silenced plants is partially dependent on NRCX paralogs NRC2 and NRC3, but not NRC4. Despite its negative impact on plant growth when silenced systemically, spot gene silencing of NRCX in mature N. benthamiana leaves doesn't result in visible cell death phenotypes. However, alteration of NRCX expression modulates the hypersensitive response mediated by NRC2 and NRC3 in a manner consistent with a negative role for NRCX in the NRC network. We conclude that NRCX is an atypical member of the NRC network that has evolved to contribute to the homeostasis of this genetically unlinked NLR network.
Copyright: © 2023 Adachi et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
I have read the journal’s policy and the authors of this manuscript have the following competing interests: S.K. receives funding from industry on NLR biology.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

